5OEY
 
 | | Crystal structure of Leishmania major fructose-1,6-bisphosphatase in holo form. | | 分子名称: | CITRIC ACID, FBP protein, MANGANESE (II) ION, ... | | 著者 | Yuan, M, Vasquez-Valdivieso, M.G, McNae, I.W, Michels, P.A.M, Fothergill-Gilmore, L.A, Walkinshaw, M.D. | | 登録日 | 2017-07-10 | | 公開日 | 2017-09-20 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Structures of Leishmania Fructose-1,6-Bisphosphatase Reveal Species-Specific Differences in the Mechanism of Allosteric Inhibition.
J. Mol. Biol., 429, 2017
|
|
8VTO
 
 | |
1P2I
 
 | | Structural consequences of accommodation of four non-cognate amino-acid residues in the S1 pocket of bovine trypsin and chymotrypsin | | 分子名称: | CALCIUM ION, Pancreatic trypsin inhibitor, SULFATE ION, ... | | 著者 | Helland, R, Czapinska, H, Leiros, I, Olufsen, M, Otlewski, J, Smalaas, A.O. | | 登録日 | 2003-04-15 | | 公開日 | 2004-04-20 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural consequences of accommodation of four non-cognate amino acid residues in the S1 pocket of bovine trypsin and chymotrypsin.
J.Mol.Biol., 333, 2003
|
|
8VTL
 
 | |
6AXK
 
 | | Crystal structure of Fab311 complex | | 分子名称: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACE-ASN-PRO-ASN-ALA-ASN-PRO-ASN-ALA-ASN-PRO-ASN, Fab311 heavy chain, ... | | 著者 | Oyen, D, Wilson, I.A. | | 登録日 | 2017-09-07 | | 公開日 | 2017-11-22 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.103 Å) | | 主引用文献 | Structural basis for antibody recognition of the NANP repeats in Plasmodium falciparum circumsporozoite protein.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8VTM
 
 | |
1ISV
 
 | | Crystal structure of xylanase from Streptomyces olivaceoviridis E-86 complexed with xylose | | 分子名称: | beta-D-xylopyranose, endo-1,4-beta-D-xylanase | | 著者 | Fujimoto, Z, Kuno, A, Kaneko, S, Kobayashi, H, Kusakabe, I, Mizuno, H. | | 登録日 | 2001-12-27 | | 公開日 | 2002-02-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structures of the sugar complexes of Streptomyces olivaceoviridis E-86 xylanase: sugar binding structure of the family 13 carbohydrate binding module.
J.Mol.Biol., 316, 2002
|
|
5OH1
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Aminoglutethimide | | 分子名称: | (3~{R})-3-(4-aminophenyl)-3-ethyl-piperidine-2,6-dione, Cereblon isoform 4, S-Thalidomide, ... | | 著者 | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | 登録日 | 2017-07-14 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
7S4Y
 
 | | Serial Macromolecular Crystallography at ALBA Synchrotron Light Source - Insulin | | 分子名称: | CHLORIDE ION, Insulin A chain, Insulin B chain, ... | | 著者 | Martin-Garcia, J.M, Botha, S, Hu, H, Jernigan, R, Castellvi, A, Lisova, S, Gil, F, Calisto, B, Crespo, I, Roy-Chowdbury, S, Grieco, A, Ketawala, G, Weierstall, U, Spence, J, Fromme, P, Zatsepin, N, Boer, R, Carpena, X. | | 登録日 | 2021-09-09 | | 公開日 | 2021-10-27 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.71 Å) | | 主引用文献 | Serial macromolecular crystallography at ALBA Synchrotron Light Source.
J.Synchrotron Radiat., 29, 2022
|
|
7RZA
 
 | | Structure of the complex of AMPA receptor GluA2 with auxiliary subunit GSG1L bound to agonist quisqualate | | 分子名称: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2 | | 著者 | Gangwar, S.P, Klykov, O.V, Yelshanskaya, M.V, Sobolevsky, A.I. | | 登録日 | 2021-08-27 | | 公開日 | 2021-10-27 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (4.26 Å) | | 主引用文献 | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
5OH9
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Thiazolidine-2,4-dione | | 分子名称: | 1,3-thiazole-2,4-dione, Cereblon isoform 4, ZINC ION | | 著者 | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | 登録日 | 2017-07-14 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
1J4K
 
 | |
7RYZ
 
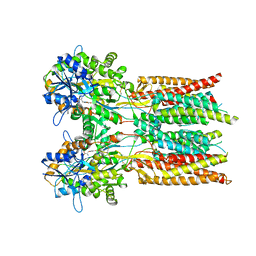 | | Structure of the complex of LBD-TMD part of AMPA receptor GluA2 with auxiliary subunit GSG1L bound to agonist quisqualate | | 分子名称: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2 | | 著者 | Gangwar, S.P, Klykov, O.V, Yelshanskaya, M.V, Sobolevsky, A.I. | | 登録日 | 2021-08-26 | | 公開日 | 2021-10-27 | | 最終更新日 | 2022-02-16 | | 実験手法 | ELECTRON MICROSCOPY (4.15 Å) | | 主引用文献 | Structure and desensitization of AMPA receptor complexes with type II TARP gamma 5 and GSG1L.
Mol.Cell, 81, 2021
|
|
2CJT
 
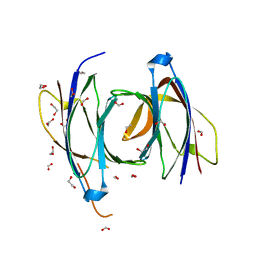 | | Structural Basis for a Munc13-1 Homodimer - Munc13-1 - RIM Heterodimer Switch: C2-domains as Versatile Protein-Protein Interaction Modules | | 分子名称: | 1,2-ETHANEDIOL, FORMIC ACID, UNC-13 HOMOLOG A | | 著者 | Lu, J, Machius, M, Dulubova, I, Dai, H, Sudhof, T.C, Tomchick, D.R, Rizo, J. | | 登録日 | 2006-04-06 | | 公開日 | 2006-06-07 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.44 Å) | | 主引用文献 | Structural Basis for a Munc13-1 Dimeric to Munc13-1/Rim Heterodimer Switch
Plos Biol., 4, 2006
|
|
6VBB
 
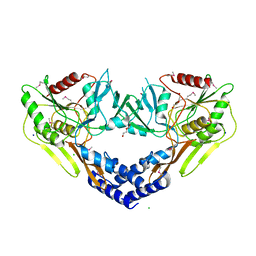 | | 2.60 Angstrom Resolution Crystal Structure of Peptidase S41 from Acinetobacter baumannii | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Peptidase S41, ... | | 著者 | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-12-18 | | 公開日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | 2.60 Angstrom Resolution Crystal Structure of Peptidase S41 from Acinetobacter baumannii
To Be Published
|
|
6VCC
 
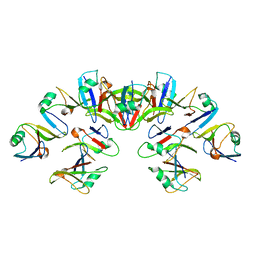 | | Cryo-EM structure of the Dvl2 DIX filament | | 分子名称: | Segment polarity protein dishevelled homolog DVL-2 | | 著者 | Enos, M, Kan, W, Muennich, S, Chen, D.H, Skiniotis, G, Weis, W.I. | | 登録日 | 2019-12-20 | | 公開日 | 2020-04-29 | | 最終更新日 | 2024-03-06 | | 実験手法 | ELECTRON MICROSCOPY (3.6 Å) | | 主引用文献 | Limited Dishevelled/Axin oligomerization determines efficiency of Wnt/ beta-catenin signal transduction.
Elife, 9, 2020
|
|
5OFQ
 
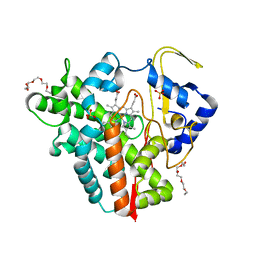 | |
8W0W
 
 | |
5OH3
 
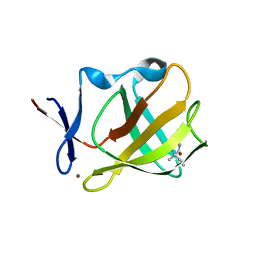 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Ethosuximide | | 分子名称: | (3~{S})-3-ethyl-3-methyl-pyrrolidine-2,5-dione, Cereblon isoform 4, ZINC ION | | 著者 | Boichenko, I, Albrecht, R, Lupas, A.N, Hernandez Alvarez, B, Hartmann, M.D. | | 登録日 | 2017-07-14 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Chemical Ligand Space of Cereblon.
Acs Omega, 3, 2018
|
|
7QR3
 
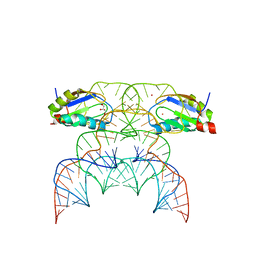 | | Chimpanzee CPEB3 HDV-like ribozyme | | 分子名称: | GLYCEROL, POTASSIUM ION, U1 small nuclear ribonucleoprotein A, ... | | 著者 | Przytula-Mally, A.I, Engilberge, S, Johannsen, S, Olieric, V, Masquida, B, Sigel, R.K.O. | | 登録日 | 2022-01-10 | | 公開日 | 2022-10-26 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Anticodon-like loop-mediated dimerization in the crystal structures of HdV-like CPEB3 ribozymes
Biorxiv, 2022
|
|
2J6F
 
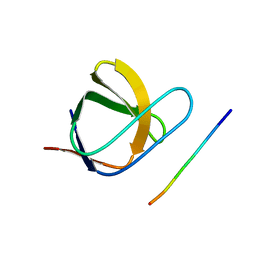 | | N-TERMINAL SH3 DOMAIN OF CMS (CD2AP HUMAN HOMOLOG) BOUND TO CBL-B PEPTIDE | | 分子名称: | CD2-ASSOCIATED PROTEIN, E3 UBIQUITIN-PROTEIN LIGASE CBL-B | | 著者 | Moncalian, G, Cardenes, N, Deribe, Y.L, Spinola-Amilibia, M, Dikic, I, Bravo, J. | | 登録日 | 2006-09-28 | | 公開日 | 2006-10-11 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Atypical Polyproline Recognition by the Cms N- Terminal SH3 Domain.
J.Biol.Chem., 281, 2006
|
|
2J8A
 
 | | X-ray structure of the N-terminus RRM domain of Set1 | | 分子名称: | HISTONE-LYSINE N-METHYLTRANSFERASE, H3 LYSINE-4 SPECIFIC | | 著者 | Tresaugues, L, Dehe, P.M, Guerois, R, Rodriguez-Gil, A, Varlet, I, Salah, P, Pamblanco, M, Luciano, P, Quevillon-Cheruel, S, Sollier, J, Leulliot, N, Couprie, J, Tordera, V, Zinn-Justin, S, Chavez, S, Van Tilbeurgh, H, Geli, V. | | 登録日 | 2006-10-24 | | 公開日 | 2007-03-20 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | X-Ray Structure of the N-Terminus Rrm Domain of Set1
To be Published
|
|
2WNN
 
 | | Structure of wild type E. coli N-acetylneuraminic acid lyase in complex with pyruvate in space group P21 | | 分子名称: | N-ACETYLNEURAMINATE LYASE, PENTAETHYLENE GLYCOL, SODIUM ION | | 著者 | Campeotto, I, Bolt, A.H, Harman, T.A, Trinh, C.H, Dennis, C.A, Phillips, S.E.V, Pearson, A.R, Nelson, A, Berry, A. | | 登録日 | 2009-07-13 | | 公開日 | 2010-08-25 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural Insights Into Substrate Specificity in Variants of N-Acetylneuraminic Acid Lyase Produced by Directed Evolution.
J.Mol.Biol., 404, 2010
|
|
6Q5V
 
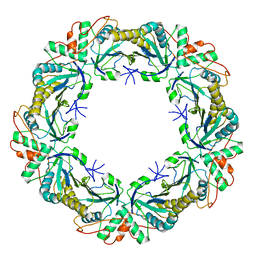 | | 1-Cys SiPrx, a Prx6-family 1-Cys peroxiredoxin of the thermoacidophilic archaeon Sulfolobus islandicus | | 分子名称: | Peroxiredoxin | | 著者 | Stroobants, S, Maes, D, Peeters, E, van Molle, I. | | 登録日 | 2018-12-09 | | 公開日 | 2019-05-29 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.747 Å) | | 主引用文献 | Structure of the Prx6-subfamily 1-Cys peroxiredoxin from Sulfolobus islandicus.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
1LC7
 
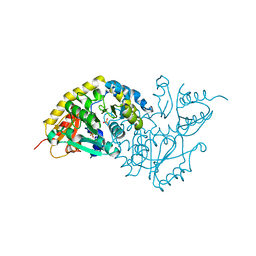 | | Crystal Structure of L-Threonine-O-3-phosphate Decarboxylase from S. enterica complexed with a substrate | | 分子名称: | L-Threonine-O-3-Phosphate Decarboxylase, PHOSPHATE ION, PHOSPHOTHREONINE | | 著者 | Cheong, C.-G, Escalante-Semerena, J, Rayment, I. | | 登録日 | 2002-04-05 | | 公開日 | 2002-06-28 | | 最終更新日 | 2023-08-16 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structural studies of the L-threonine-O-3-phosphate decarboxylase (CobD) enzyme from Salmonella enterica: the apo, substrate, and product-aldimine complexes.
Biochemistry, 41, 2002
|
|
