7SYH
 
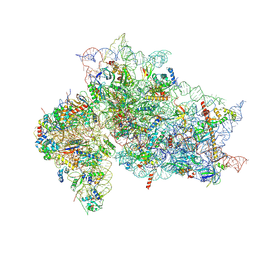 | | Structure of the HCV IRES binding to the 40S ribosomal subunit, closed conformation. Structure 2(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S21, 40S ribosomal protein S24, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-13 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Molecular architecture of 40S initiation complexes on the Hepatitis C virus IRES: from ribosomal attachment to eIF5B-mediated reorientation of initiator tRNA
To Be Published
|
|
8V2E
 
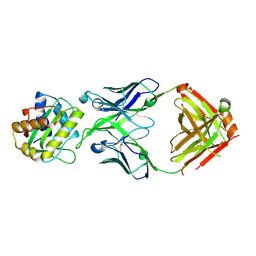 | |
7SNM
 
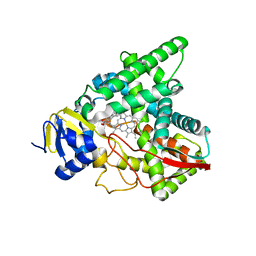 | |
3EJD
 
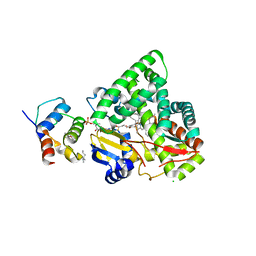 | |
7SYP
 
 | | Structure of the wt IRES and 40S ribosome binary complex, open conformation. Structure 10(wt) | | 分子名称: | 18S rRNA, HCV IRES, HCV IRES partially loaded mRNA portion, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-27 | | 最終更新日 | 2022-08-24 | | 実験手法 | ELECTRON MICROSCOPY (4 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
7SYO
 
 | | Structure of the HCV IRES bound to the 40S ribosomal subunit, head open. Structure 9(delta dII) | | 分子名称: | 18S rRNA, 40S ribosomal protein S2, HCV IRES, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-27 | | 最終更新日 | 2022-08-24 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
6TPA
 
 | | CDK8/CyclinC in complex with drug ETP-50775 | | 分子名称: | (2~{R})-butane-1,2-diol, 1-[4-chloranyl-3-(trifluoromethyl)phenyl]-3-(5-oxidanylidene-6-pyridin-4-yl-pyrido[2,3-b][1,5]benzoxazepin-9-yl)urea, Cyclin-C, ... | | 著者 | Munoz, I.G, Pastor, J, Martinez, S. | | 登録日 | 2019-12-12 | | 公開日 | 2020-11-18 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Pyrido[2,3-b][1,5]benzoxazepin-5(6H)-one derivatives as CDK8 inhibitors.
Eur.J.Med.Chem., 201, 2020
|
|
4UCF
 
 | | Crystal structure of Bifidobacterium bifidum beta-galactosidase in complex with alpha-galactose | | 分子名称: | BETA-GALACTOSIDASE, DI(HYDROXYETHYL)ETHER, N-PROPANOL, ... | | 著者 | Godoy, A.S, Murakami, M.T, Camilo, C.M, Bernardes, A, Polikarpov, I. | | 登録日 | 2014-12-03 | | 公開日 | 2016-01-20 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.94 Å) | | 主引用文献 | Crystal Structure of Beta1-6-Galactosidase from Bifidobacterium Bifidum S17: Trimeric Architecture, Molecular Determinants of the Enzymatic Activity and its Inhibition by Alpha-Galactose.
FEBS J., 283, 2016
|
|
7SYQ
 
 | | Structure of the wt IRES and 40S ribosome ternary complex, open conformation. Structure 11(wt) | | 分子名称: | 18S rRNA, Eukaryotic translation initiation factor 1A, X-chromosomal, ... | | 著者 | Brown, Z.P, Abaeva, I.S, De, S, Hellen, C.U.T, Pestova, T.V, Frank, J. | | 登録日 | 2021-11-25 | | 公開日 | 2022-07-27 | | 最終更新日 | 2022-08-24 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Molecular architecture of 40S translation initiation complexes on the hepatitis C virus IRES.
Embo J., 41, 2022
|
|
8H2C
 
 | | Crystal structure of the pseudaminic acid synthase PseI from Campylobacter jejuni | | 分子名称: | MANGANESE (II) ION, Pseudaminic acid synthase | | 著者 | Song, W.S, Park, M.A, Ki, D.U, Yoon, S.I. | | 登録日 | 2022-10-05 | | 公開日 | 2022-11-09 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structural analysis of the pseudaminic acid synthase PseI from Campylobacter jejuni.
Biochem.Biophys.Res.Commun., 635, 2022
|
|
7SOK
 
 | | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with inhibitor II329 | | 分子名称: | (2S)-2-amino-4-([3-(3-carbamoylphenyl)prop-2-yn-1-yl]{[(1R,2R,3S,4R)-4-(4-chloro-7H-pyrrolo[2,3-d]pyrimidin-7-yl)-2,3-dihydroxycyclopentyl]methyl}amino)butanoic acid, DI(HYDROXYETHYL)ETHER, NNMT protein | | 著者 | Yadav, R, Iyamu, I.D, Huang, R, Noinaj, N. | | 登録日 | 2021-10-31 | | 公開日 | 2022-11-02 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.08 Å) | | 主引用文献 | Structure of Nicotinamide N-Methyltransferase (NNMT) in complex with inhibitor II329
To Be Published
|
|
8H52
 
 | | Crystal structure of Helicobacter pylori carboxyspermidine dehydrogenase in complex with NADP | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Saccharopine dehydrogenase | | 著者 | Ko, K.Y, Park, S.C, Cho, S.Y, Yoon, S.I. | | 登録日 | 2022-10-11 | | 公開日 | 2022-11-09 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural analysis of carboxyspermidine dehydrogenase from Helicobacter pylori.
Biochem.Biophys.Res.Commun., 635, 2022
|
|
7T7B
 
 | |
8H2I
 
 | | Near-atomic structure of five-fold averaged PBCV-1 capsid | | 分子名称: | MCPv1, MCPv2, MCPv3, ... | | 著者 | Shao, Q, Agarkova, I.V, Noel, E.A, Dunigan, D.D, Liu, Y, Wang, A, Guo, M, Xie, L, Zhao, X, Rossmann, M.G, Van Etten, J.L, Klose, T, Fang, Q. | | 登録日 | 2022-10-06 | | 公開日 | 2022-11-16 | | 最終更新日 | 2023-09-06 | | 実験手法 | ELECTRON MICROSCOPY (3.8 Å) | | 主引用文献 | Near-atomic, non-icosahedrally averaged structure of giant virus Paramecium bursaria chlorella virus 1.
Nat Commun, 13, 2022
|
|
1E27
 
 | | Nonstandard peptide binding of HLA-B*5101 complexed with HIV immunodominant epitope KM1(LPPVVAKEI) | | 分子名称: | BETA-2 MICROGLOBULIN LIGHT CHAIN, HIV-1 PEPTIDE (LPPVVAKEI), HLA CLASS I HISTOCOMPATIBILITY ANTIGEN HEAVY CHAIN | | 著者 | Maenaka, K, Maenaka, T, Tomiyama, H, Takiguchi, M, Stuart, D.I, Jones, E.Y. | | 登録日 | 2000-05-18 | | 公開日 | 2000-09-12 | | 最終更新日 | 2024-05-01 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Nonstandard peptide binding revealed by crystal structures of HLA-B*5101 complexed with HIV immunodominant epitopes.
J Immunol., 165, 2000
|
|
5S4I
 
 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF051 | | 分子名称: | (5S)-1-(4-chlorophenyl)-5-methylimidazolidine-2,4-dione, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.131 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
4U4N
 
 | | Crystal structure of Edeine bound to the yeast 80S ribosome | | 分子名称: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | 著者 | Garreau de Loubresse, N, Prokhorova, I, Yusupova, G, Yusupov, M. | | 登録日 | 2014-07-24 | | 公開日 | 2014-10-22 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (3.1 Å) | | 主引用文献 | Structural basis for the inhibition of the eukaryotic ribosome.
Nature, 513, 2014
|
|
3E77
 
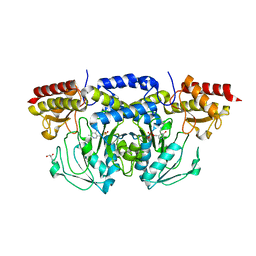 | | Human phosphoserine aminotransferase in complex with PLP | | 分子名称: | GLYCEROL, PYRIDOXAL-5'-PHOSPHATE, Phosphoserine aminotransferase | | 著者 | Lehtio, L, Karlberg, T, Andersson, J, Arrowsmith, C.H, Berglund, H, Bountra, C, Collins, R, Dahlgren, L.G, Edwards, A.M, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Johansson, A, Johansson, I, Kotenyova, T, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Olesen, K, Persson, C, Sagemark, J, Thorsell, S.G, Tresaugues, L, Van Den Berg, S, Welin, M, Wikstrom, M, Wisniewska, M, Weigelt, J, Schueler, H, Structural Genomics Consortium (SGC) | | 登録日 | 2008-08-18 | | 公開日 | 2008-08-26 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Human phosphoserine aminotransferase in complex with PLP
TO BE PUBLISHED
|
|
5S4H
 
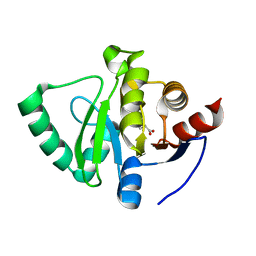 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF048 | | 分子名称: | 1-carbamoylpiperidine-4-carboxylic acid, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.175 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
3EJB
 
 | |
5S4F
 
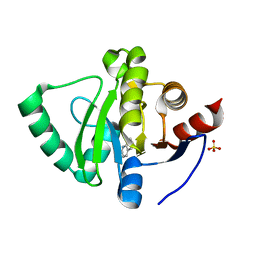 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with SF003 | | 分子名称: | 1,8-naphthyridine, Non-structural protein 3, SULFATE ION | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.131 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
7NE7
 
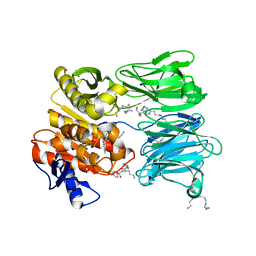 | | oligopeptidase B from S. proteomaculans with modified hinge region in complex with N-[(1S)-5-amino-1-(chloroacetyl)pentyl]-4-methylbenzenesulfonamide | | 分子名称: | N-[(1S)-5-amino-1-(chloroacetyl)pentyl]-4-methylbenzenesulfonamide, Oligopeptidase B, SPERMINE | | 著者 | Petrenko, D.E, Nikolaeva, A.Y, Lazarenko, V.A, Dorovatovskiy, P.V, Vlaskina, A.V, Mikhailova, A.G, Rakitina, T.V, Timofeev, V.I. | | 登録日 | 2021-02-03 | | 公開日 | 2021-02-17 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | The Crystal Structure of N alpha-p-tosyl-lysyl Chloromethylketone-Bound Oligopeptidase B from Serratia Proteamaculans Revealed a New Type of Inhibitor Binding
Crystals, 11, 2021
|
|
7AER
 
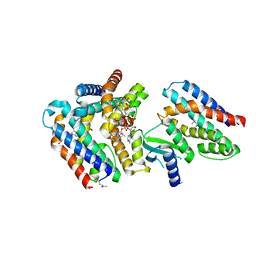 | | Rebuilt and re-refined PDB entry 5yep: tri-AMPylated Shewanella oneidensis HEPN toxin in complex with MNT antitoxin | | 分子名称: | ADENOSINE MONOPHOSPHATE, Toxin-antitoxin system antidote Mnt family, Toxin-antitoxin system toxin HepN family | | 著者 | Tamulaitiene, G, Sasnauskas, G, Songailiene, I, Juozapaitis, J, Siksnys, V. | | 登録日 | 2020-09-18 | | 公開日 | 2020-12-30 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | HEPN-MNT Toxin-Antitoxin System: The HEPN Ribonuclease Is Neutralized by OligoAMPylation.
Mol.Cell, 80, 2020
|
|
5OXZ
 
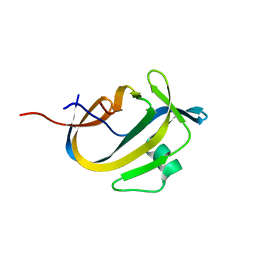 | | Crystal Structure of NeqN/NeqC complex from Nanoarcheaum equitans at 1.2A | | 分子名称: | NEQ068, NEQ528 | | 著者 | Aparicio, D, Perez-Luque, R, Ribo, M, Fita, I. | | 登録日 | 2017-09-07 | | 公開日 | 2018-10-10 | | 最終更新日 | 2024-05-08 | | 実験手法 | X-RAY DIFFRACTION (1.2 Å) | | 主引用文献 | Structural Insights into Subunits Assembly and the Oxyester Splicing Mechanism of Neq pol Split Intein.
Cell Chem Biol, 25, 2018
|
|
5S40
 
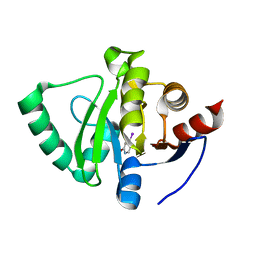 | | PanDDA analysis group deposition -- Crystal Structure of SARS-CoV-2 Nsp3 macrodomain in complex with NCL-00023824 | | 分子名称: | 4-iodanyl-3~{H}-pyridin-2-one, Non-structural protein 3 | | 著者 | Fearon, D, Schuller, M, Rangel, V.L, Douangamath, A, Rack, J.G.M, Zhu, K, Aimon, A, Brandao-Neto, J, Dias, A, Dunnet, L, Gorrie-Stone, T.J, Powell, A.J, Krojer, T, Skyner, R, Thompson, W, Ahel, I, von Delft, F. | | 登録日 | 2020-11-02 | | 公開日 | 2021-01-13 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.187 Å) | | 主引用文献 | Fragment binding to the Nsp3 macrodomain of SARS-CoV-2 identified through crystallographic screening and computational docking.
Sci Adv, 7, 2021
|
|
