6M13
 
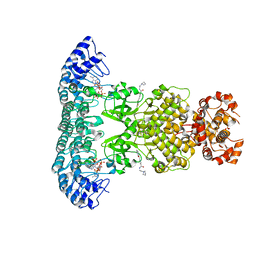 | | Crystal structure of Rnase L in complex with Toceranib | | 分子名称: | 5-[(Z)-(5-fluoranyl-2-oxidanylidene-1H-indol-3-ylidene)methyl]-2,4-dimethyl-N-(2-pyrrolidin-1-ylethyl)-1H-pyrrole-3-carboxamide, PHOSPHATE ION, Ribonuclease L, ... | | 著者 | Tang, J, Huang, H. | | 登録日 | 2020-02-24 | | 公開日 | 2020-09-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.56 Å) | | 主引用文献 | Sunitinib inhibits RNase L by destabilizing its active dimer conformation.
Biochem.J., 477, 2020
|
|
5UHW
 
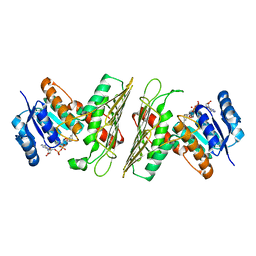 | | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+ and Magnesium | | 分子名称: | MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Oxidoreductase protein | | 著者 | Cook, W.J, Fedorov, A.A, Fedorov, E.V, Huang, H, Bonanno, J.B, Gerlt, J.A, Almo, S.C. | | 登録日 | 2017-01-12 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.24 Å) | | 主引用文献 | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+ and Magnesium
To be published
|
|
5UHZ
 
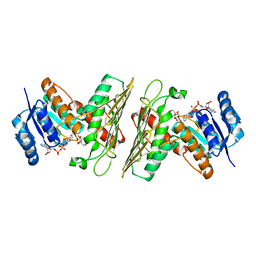 | | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+, D-Apionate and Magnesium | | 分子名称: | (3R,4R)-3,4-dihydroxy-4-(hydroxymethyl)oxolan-2-one, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Cook, W.J, Fedorov, A.A, Fedorov, E.V, Huang, H, Bonanno, J.B, Gerlt, J.A, Almo, S.C. | | 登録日 | 2017-01-12 | | 公開日 | 2017-02-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+, D-Apionate and Magnesium
To be published
|
|
5UIB
 
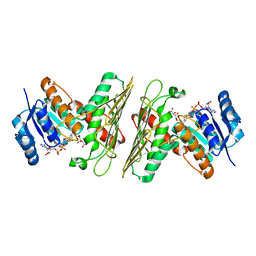 | | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+, L-tartaric acid and Magnesium | | 分子名称: | L(+)-TARTARIC ACID, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Cook, W.J, Fedorov, A.A, Fedorov, E.V, Huang, H, Bonanno, J.B, Gerlt, J.A, Almo, S.C. | | 登録日 | 2017-01-13 | | 公開日 | 2017-01-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+, L-tartaric acid and Magnesium
To be published
|
|
5UIA
 
 | | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+, R-2,3-dihydroxyisovalerate and Magnesium | | 分子名称: | (2S)-2,3-dihydroxy-3-methylbutanoic acid, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Cook, W.J, Fedorov, A.A, Fedorov, E.V, Huang, H, Bonanno, J.B, Gerlt, J.A, Almo, S.C. | | 登録日 | 2017-01-13 | | 公開日 | 2017-02-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.18 Å) | | 主引用文献 | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+, R-2,3-dihydroxyisovalerate and Magnesium
To be published
|
|
5T57
 
 | | Crystal Structure of a Semialdehyde dehydrogenase NAD-binding Protein from Cupriavidus necator in Complex with Calcium and NAD | | 分子名称: | CALCIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, Semialdehyde dehydrogenase NAD-binding protein, ... | | 著者 | Cook, W.J, Fedorov, A.A, Fedorov, E.V, Huang, H, Bonanno, J.B, Gerlt, J.A, Almo, S.C. | | 登録日 | 2016-08-30 | | 公開日 | 2016-09-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Crystal Structure of a Semialdehyde dehydrogenase NAD-binding Protein from Cupriavidus necator in Complex with Calcium and NAD
To be published
|
|
3GYP
 
 | | Rtt106p | | 分子名称: | BETA-MERCAPTOETHANOL, CHLORIDE ION, Histone chaperone RTT106 | | 著者 | Liu, Y, Huang, H, Shi, Y, Teng, M. | | 登録日 | 2009-04-04 | | 公開日 | 2009-12-08 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (2.406 Å) | | 主引用文献 | Structural analysis of Rtt106p reveals a DNA-binding role required for heterochromatin silencing
J.Biol.Chem., 285, 2010
|
|
5TNV
 
 | | Crystal Structure of a Xylose isomerase-like TIM barrel Protein from Mycobacterium smegmatis in Complex with Magnesium | | 分子名称: | AP endonuclease, family protein 2, MAGNESIUM ION | | 著者 | Cook, W.J, Fedorov, A.A, Fedorov, E.V, Huang, H, Bonanno, J.B, Gerlt, J.A, Almo, S.C. | | 登録日 | 2016-10-14 | | 公開日 | 2016-11-09 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.04 Å) | | 主引用文献 | Crystal Structure of a Xylose isomerase-like TIM barrel Protein from Mycobacterium smegmatis in Complex with Magnesium
To Be Published
|
|
5VEI
 
 | | Crystal structure of the SH3 domain of human sorbin and SH3 domain-containing protein 2 | | 分子名称: | Sorbin and SH3 domain-containing protein 2, UNKNOWN ATOM OR ION | | 著者 | Liu, Y, Tempel, W, Huang, H, Gu, J, Liu, K, Sidhu, S.S, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | 登録日 | 2017-04-04 | | 公開日 | 2017-08-02 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.33 Å) | | 主引用文献 | Crystal structure of the SH3 domain of human sorbin and SH3 domain-containing protein 2
To be Published
|
|
5UI9
 
 | | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+, 2 -hydroxy-2-hydroxymethyl propanoic acid and Magnesium | | 分子名称: | (2S)-2,3-dihydroxy-2-methylpropanoic acid, MAGNESIUM ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | 著者 | Cook, W.J, Fedorov, A.A, Fedorov, E.V, Huang, H, Bonanno, J.B, Gerlt, J.A, Almo, S.C. | | 登録日 | 2017-01-13 | | 公開日 | 2017-02-01 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.92 Å) | | 主引用文献 | Crystal Structure of an Oxidoreductase from Agrobacterium radiobacter in Complex with NAD+, 2 -hydroxy-2-hydroxymethyl propanoic acid and Magnesium
To be published
|
|
5VR7
 
 | | ABA-mimicking ligand AMF1alpha in complex with ABA receptor PYL2 and PP2C HAB1 | | 分子名称: | 1-(2-fluoro-4-methylphenyl)-N-(2-oxo-1-propyl-1,2,3,4-tetrahydroquinolin-6-yl)methanesulfonamide, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | 著者 | Cao, M.-J, Zhang, Y.-L, Liu, X, Huang, H, Zhou, X.E, Wang, W.-L, Zeng, A, Zhao, C.-Z, Si, T, Du, J.-M, Wu, W.-W, Wang, F.-X, Xu, H.E, Zhu, J.-K. | | 登録日 | 2017-05-10 | | 公開日 | 2017-11-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.612 Å) | | 主引用文献 | Combining chemical and genetic approaches to increase drought resistance in plants.
Nat Commun, 8, 2017
|
|
5VS5
 
 | | ABA-mimicking ligand AMF2alpha in complex with ABA receptor PYL2 and PP2C HAB1 | | 分子名称: | 1-(2,3-difluoro-4-methylphenyl)-N-(2-oxo-1-propyl-1,2,3,4-tetrahydroquinolin-6-yl)methanesulfonamide, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | 著者 | Cao, M.-J, Zhang, Y.-L, Liu, X, Huang, H, Zhou, X.E, Wang, W.-W, Zeng, A, Zhao, C.-Z, Si, T, Du, J.-M, Wu, W.-W, Wang, F.-X, Xu, H.E, Zhu, J.-K. | | 登録日 | 2017-05-11 | | 公開日 | 2017-11-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Combining chemical and genetic approaches to increase drought resistance in plants.
Nat Commun, 8, 2017
|
|
5VSQ
 
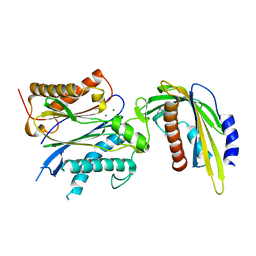 | | ABA-mimicking ligand AMF2beta in complex with ABA receptor PYL2 and PP2C HAB1 | | 分子名称: | 1-(3,5-difluoro-4-methylphenyl)-N-(2-oxo-1-propyl-1,2,3,4-tetrahydroquinolin-6-yl)methanesulfonamide, Abscisic acid receptor PYL2, MAGNESIUM ION, ... | | 著者 | Cao, M.-J, Zhang, Y.-L, Liu, X, Huang, H, Zhou, X.E, Wang, W.-L, Zeng, A, Zhao, C.-Z, Si, T, Du, J.-M, Wu, W.-W, Wang, F.-X, Xu, H.X, Zhu, J.-K. | | 登録日 | 2017-05-12 | | 公開日 | 2017-11-15 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.618 Å) | | 主引用文献 | Combining chemical and genetic approaches to increase drought resistance in plants.
Nat Commun, 8, 2017
|
|
3R45
 
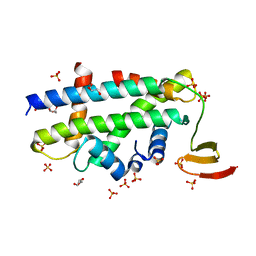 | | Structure of a CENP-A-Histone H4 Heterodimer in complex with chaperone HJURP | | 分子名称: | GLYCEROL, Histone H3-like centromeric protein A, Histone H4, ... | | 著者 | Hu, H, Liu, Y, Wang, M, Fang, J, Huang, H, Yang, N, Li, Y, Wang, J, Yao, X, Shi, Y, Li, G, Xu, R.M. | | 登録日 | 2011-03-17 | | 公開日 | 2011-04-06 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structure of a CENP-A-histone H4 heterodimer in complex with chaperone HJURP
Genes Dev., 25, 2011
|
|
3DEA
 
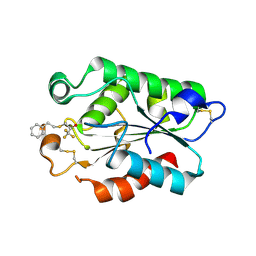 | | Glomerella cingulata PETFP-cutinase complex | | 分子名称: | 1,1,1-trifluoro-3-[(2-phenylethyl)sulfanyl]propan-2-one, Cutinase | | 著者 | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | 登録日 | 2008-06-09 | | 公開日 | 2008-11-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
1DDP
 
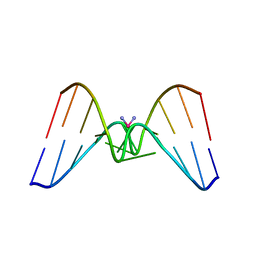 | | Solution structure of a CISPLATIN-INDUCED [CATAGCTATG]2 Interstrand cross-link | | 分子名称: | Cisplatin, DNA (5'-D(*CP*AP*TP*AP*GP*CP*TP*AP*TP*G)-3') | | 著者 | Zhu, L, Huang, H, Reid, B.R, Drobny, G.P, Hopkins, P.B. | | 登録日 | 1995-10-26 | | 公開日 | 1996-03-08 | | 最終更新日 | 2024-03-13 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of a cisplatin-induced DNA interstrand cross-link.
Science, 270, 1995
|
|
1RTD
 
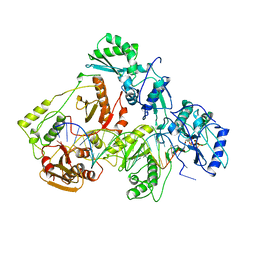 | | STRUCTURE OF A CATALYTIC COMPLEX OF HIV-1 REVERSE TRANSCRIPTASE: IMPLICATIONS FOR NUCLEOSIDE ANALOG DRUG RESISTANCE | | 分子名称: | DNA PRIMER FOR REVERSE TRANSCRIPTASE, DNA TEMPLATE FOR REVERSE TRANSCRIPTASE, MAGNESIUM ION, ... | | 著者 | Chopra, R, Huang, H, Verdine, G.L, Harrison, S.C. | | 登録日 | 1998-08-26 | | 公開日 | 1998-12-09 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance.
Science, 282, 1998
|
|
4R4P
 
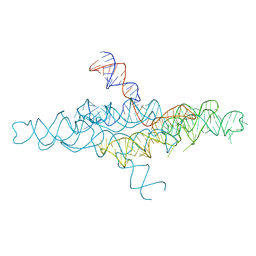 | | Crystal Structure of the VS ribozyme-A756G mutant | | 分子名称: | MAGNESIUM ION, VS ribozyme RNA | | 著者 | Piccirilli, J.A, Suslov, N.B, Dasgupta, S, Huang, H, Lilley, D.M.J, Rice, P.A. | | 登録日 | 2014-08-19 | | 公開日 | 2015-09-30 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | Crystal structure of the Varkud satellite ribozyme.
Nat.Chem.Biol., 11, 2015
|
|
4R4V
 
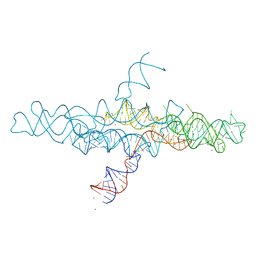 | | Crystal structure of the VS ribozyme - G638A mutant | | 分子名称: | MAGNESIUM ION, POTASSIUM ION, VS ribozyme RNA | | 著者 | Piccirilli, J.A, Suslov, N.B, Dasgupta, S, Huang, H, Lilley, D.M.J, Rice, P.A. | | 登録日 | 2014-08-19 | | 公開日 | 2015-09-30 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (3.07 Å) | | 主引用文献 | Crystal structure of the Varkud satellite ribozyme.
Nat.Chem.Biol., 11, 2015
|
|
1F3H
 
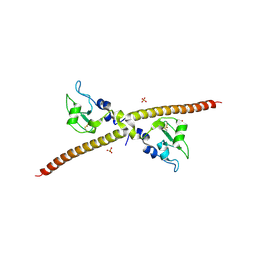 | | X-RAY CRYSTAL STRUCTURE OF THE HUMAN ANTI-APOPTOTIC PROTEIN SURVIVIN | | 分子名称: | SULFATE ION, SURVIVIN, ZINC ION | | 著者 | Verdecia, M.A, Huang, H, Dutil, E, Hunter, T, Noel, J.P. | | 登録日 | 2000-06-03 | | 公開日 | 2000-12-06 | | 最終更新日 | 2021-11-03 | | 実験手法 | X-RAY DIFFRACTION (2.58 Å) | | 主引用文献 | Structure of the human anti-apoptotic protein survivin reveals a dimeric arrangement.
Nat.Struct.Biol., 7, 2000
|
|
3DD5
 
 | | Glomerella cingulata E600-cutinase complex | | 分子名称: | Cutinase, DIETHYL PHOSPHONATE | | 著者 | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | 登録日 | 2008-06-05 | | 公開日 | 2008-11-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
3DCN
 
 | | Glomerella cingulata apo cutinase | | 分子名称: | Cutinase | | 著者 | Nyon, M.P, Rice, D.W, Berrisford, J.M, Hounslow, A.M, Moir, A.J.G, Huang, H, Nathan, S, Mahadi, N.M, Farah Diba, A.B, Craven, C.J. | | 登録日 | 2008-06-04 | | 公開日 | 2008-11-18 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Catalysis by Glomerella cingulata Cutinase Requires Conformational Cycling between the Active and Inactive States of Its Catalytic Triad
J.Mol.Biol., 385, 2009
|
|
6LOJ
 
 | | The complex structure of IpaH9.8-LRR and hGBP1 | | 分子名称: | GUANOSINE-5'-DIPHOSPHATE, Guanylate-binding protein 1, Invasion plasmid antigen | | 著者 | Ye, Y, Huang, H. | | 登録日 | 2020-01-06 | | 公開日 | 2020-12-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (3.72 Å) | | 主引用文献 | Substrate-binding destabilizes the hydrophobic cluster to relieve the autoinhibition of bacterial ubiquitin ligase IpaH9.8.
Commun Biol, 3, 2020
|
|
6LOL
 
 | | The crystal structure of full length IpaH9.8 | | 分子名称: | E3 ubiquitin-protein ligase ipaH9.8 | | 著者 | Ye, Y, Huang, H. | | 登録日 | 2020-01-06 | | 公開日 | 2020-12-23 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Substrate-binding destabilizes the hydrophobic cluster to relieve the autoinhibition of bacterial ubiquitin ligase IpaH9.8.
Commun Biol, 3, 2020
|
|
6B2E
 
 | | Structure of full length human AMPK (a2b2g1) in complex with a small molecule activator SC4. | | 分子名称: | 5'-AMP-activated protein kinase catalytic subunit alpha-2, 5'-AMP-activated protein kinase subunit beta-2, 5'-AMP-activated protein kinase subunit gamma-1, ... | | 著者 | Ngoei, K.R.W, Langendorf, C.G, Ling, N.X.Y, Hoque, A, Johnson, S, Camerino, M.C, Walker, S.R, Bozikis, Y.E, Dite, T.A, Ovens, A.J, Smiles, W.J, Jacobs, R, Huang, H, Parker, M.W, Scott, J.W, Rider, M.H, Kemp, B.E, Foitzik, R.C, Baell, J.B, Oakhill, J.S. | | 登録日 | 2017-09-19 | | 公開日 | 2018-04-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.8 Å) | | 主引用文献 | Structural Determinants for Small-Molecule Activation of Skeletal Muscle AMPK alpha 2 beta 2 gamma 1 by the Glucose Importagog SC4.
Cell Chem Biol, 25, 2018
|
|
