6TF2
 
 | |
4C40
 
 | |
4C4W
 
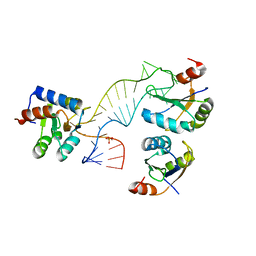 | | Structure of a rare, non-standard sequence k-turn bound by L7Ae protein | | 分子名称: | 50S RIBOSOMAL PROTEIN L7AE, DIHYDROGENPHOSPHATE ION, TSKT-23, ... | | 著者 | Huang, L, Lilley, D.M.J. | | 登録日 | 2013-09-09 | | 公開日 | 2014-02-12 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.95 Å) | | 主引用文献 | Structure of a Rare, Non-Standard Sequence K-Turn Bound by L7Ae Protein
Nucleic Acids Res., 42, 2014
|
|
4BW0
 
 | |
6TF1
 
 | |
6TFF
 
 | |
6TF3
 
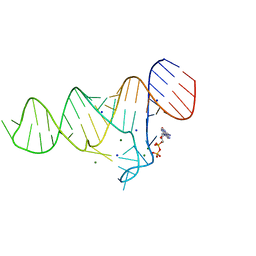 | |
4CS1
 
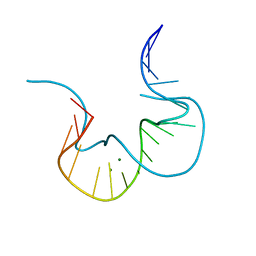 | |
3SUH
 
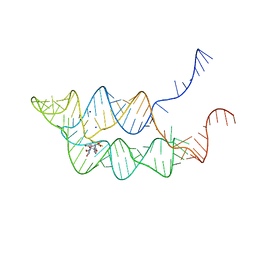 | | Crystal structure of THF riboswitch, bound with 5-formyl-THF | | 分子名称: | N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, Riboswitch, SODIUM ION | | 著者 | Huang, L, Serganov, A, Patel, D.J. | | 登録日 | 2011-07-11 | | 公開日 | 2011-09-14 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Long-range pseudoknot interactions dictate the regulatory response in the tetrahydrofolate riboswitch.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
5FJ4
 
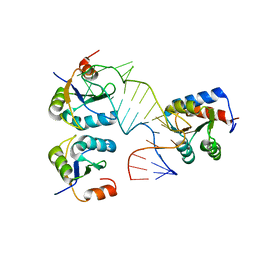 | |
5FJ0
 
 | |
5EGC
 
 | |
5G4V
 
 | |
7Y1C
 
 | |
3SUX
 
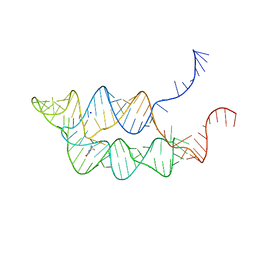 | | Crystal structure of THF riboswitch, bound with THF | | 分子名称: | 5-HYDROXYMETHYLENE-6-HYDROFOLIC ACID, Riboswitch, SODIUM ION | | 著者 | Huang, L, Serganov, A, Patel, D.J. | | 登録日 | 2011-07-11 | | 公開日 | 2011-09-14 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Long-range pseudoknot interactions dictate the regulatory response in the tetrahydrofolate riboswitch.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3SUY
 
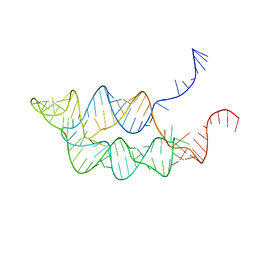 | |
7XY1
 
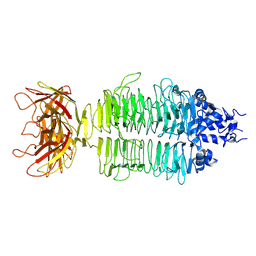 | |
7XYC
 
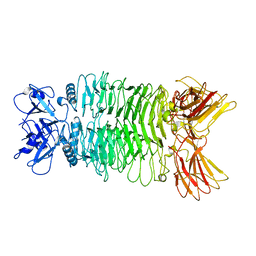 | |
7Y3T
 
 | |
7Y22
 
 | |
7Y23
 
 | |
7Y5S
 
 | |
7EAG
 
 | | Crystal structure of the RAGATH-18 k-turn | | 分子名称: | RNA (5'-R(*GP*UP*CP*UP*AP*UP*GP*AP*AP*GP*GP*CP*UP*GP*GP*AP*GP*AP*C)-3') | | 著者 | Huang, L, Lilley, D.M.J. | | 登録日 | 2021-03-07 | | 公開日 | 2021-06-02 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structure and folding of four putative kink turns identified in structured RNA species in a test of structural prediction rules.
Nucleic Acids Res., 49, 2021
|
|
7EAF
 
 | |
8WKJ
 
 | |
