5FBK
 
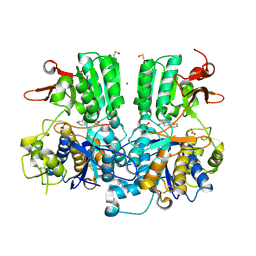 | | Crystal structure of the extracellular domain of human calcium sensing receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BICARBONATE ION, CHLORIDE ION, ... | | Authors: | Zhang, T, Zhang, C, Miller, C.L, Zou, J, Moremen, K.W, Brown, E.M, Yang, J.J, Hu, J. | | Deposit date: | 2015-12-14 | | Release date: | 2016-06-22 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for regulation of human calcium-sensing receptor by magnesium ions and an unexpected tryptophan derivative co-agonist.
Sci Adv, 2, 2016
|
|
4O2W
 
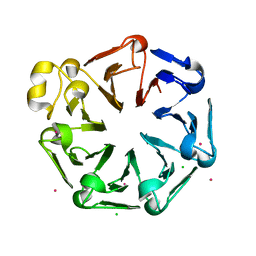 | | Crystal structure of the third RCC1-like domain of HERC1 | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase HERC1, MAGNESIUM ION, ... | | Authors: | Dong, A, Hu, J, Li, Y, Walker, J.R, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-12-17 | | Release date: | 2014-01-15 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the third RCC1-like domain of HERC1
To be Published
|
|
8EXS
 
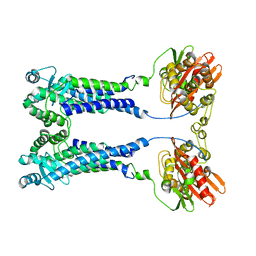 | | Cryo-EM structure of S. aureus BlaR1 F284A mutant | | Descriptor: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | Authors: | Alexander, J.A.N, Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
8EXT
 
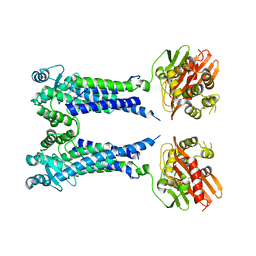 | | Cryo-EM structure of S. aureus BlaR1 F284A mutant in complex with ampicillin | | Descriptor: | Beta-lactam sensor/signal transducer BlaR1, ZINC ION | | Authors: | Alexander, J.A.N, Hu, J, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2022-10-25 | | Release date: | 2023-01-11 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis of broad-spectrum beta-lactam resistance in Staphylococcus aureus.
Nature, 613, 2023
|
|
4DAJ
 
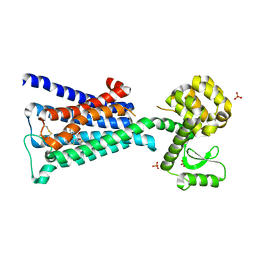 | | Structure of the M3 Muscarinic Acetylcholine Receptor | | Descriptor: | (1R,2R,4S,5S,7S)-7-{[hydroxy(dithiophen-2-yl)acetyl]oxy}-9,9-dimethyl-3-oxa-9-azoniatricyclo[3.3.1.0~2,4~]nonane, Muscarinic acetylcholine receptor M3, Lysozyme, ... | | Authors: | Kruse, A.C, Hu, J, Pan, A.C, Arlow, D.H, Rosenbaum, D.M, Rosemond, E, Green, H.F, Liu, T, Chae, P.S, Dror, R.O, Shaw, D.E, Weis, W.I, Wess, J, Kobilka, B. | | Deposit date: | 2012-01-12 | | Release date: | 2012-02-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structure and dynamics of the M3 muscarinic acetylcholine receptor.
Nature, 482, 2012
|
|
4KFW
 
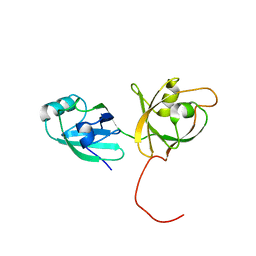 | |
4KFV
 
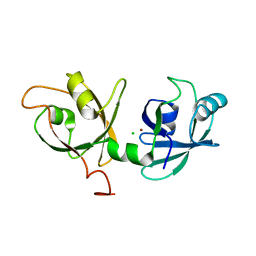 | |
5HUQ
 
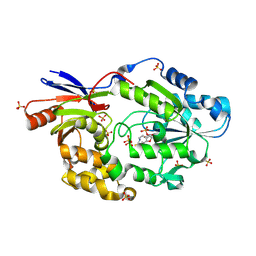 | | A tethered niacin-derived pincer complex with a nickel-carbon bond in lactate racemase | | Descriptor: | 3-methanethioyl-1-(5-O-phosphono-beta-D-ribofuranosyl)-5-(sulfanylcarbonyl)pyridin-1-ium, Lactate racemization operon protein LarA, NICKEL (II) ION, ... | | Authors: | Desguin, B, Zhang, T, Soumillion, P, Hols, P, Hu, J, Hausinger, R.P. | | Deposit date: | 2016-01-27 | | Release date: | 2016-02-10 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | METALLOPROTEINS. A tethered niacin-derived pincer complex with a nickel-carbon bond in lactate racemase.
Science, 349, 2015
|
|
4JUY
 
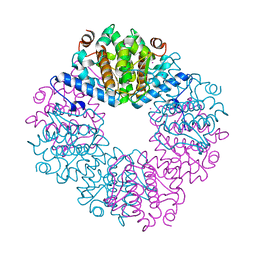 | | Crystal structure of the PUB domain of E3 ubiquitin ligase RNF31 | | Descriptor: | E3 ubiquitin-protein ligase RNF31, UNKNOWN ATOM OR ION | | Authors: | Dong, A, Hu, J, Li, Y, Wernimont, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-03-25 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the PUB domain of E3 ubiquitin ligase RNF31
To be Published
|
|
4MVT
 
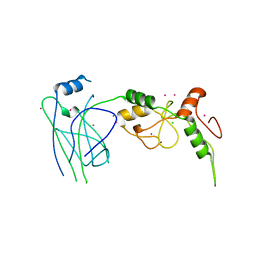 | | Crystal structure of SUMO E3 Ligase PIAS3 | | Descriptor: | CHLORIDE ION, E3 SUMO-protein ligase PIAS3, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Hu, J, Li, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2013-09-24 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of SUMO E3 Ligase PIAS3
to be published
|
|
3PMR
 
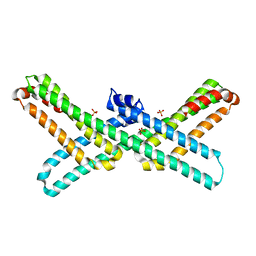 | | Crystal Structure of E2 domain of Human Amyloid Precursor-Like Protein 1 | | Descriptor: | Amyloid-like protein 1, PHOSPHATE ION | | Authors: | Lee, S, Xue, Y, Hu, J, Wang, Y, Liu, X, Demeler, B, Ha, Y. | | Deposit date: | 2010-11-17 | | Release date: | 2011-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | The E2 Domains of APP and APLP1 Share a Conserved Mode of Dimerization.
Biochemistry, 50, 2011
|
|
3NYJ
 
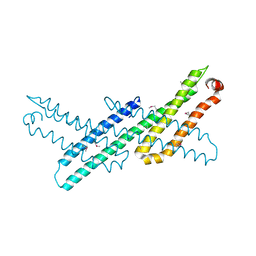 | | Crystal Structure Analysis of APP E2 domain | | Descriptor: | Amyloid beta A4 protein, OSMIUM ION | | Authors: | Ha, Y, Hu, J, Lee, S, Liu, X. | | Deposit date: | 2010-07-15 | | Release date: | 2011-06-01 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The E2 Domains of APP and APLP1 Share a Conserved Mode of Dimerization.
Biochemistry, 50, 2011
|
|
2K3K
 
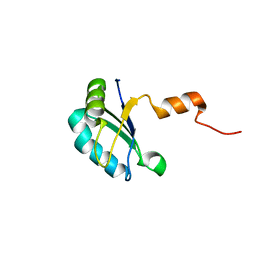 | |
5GNR
 
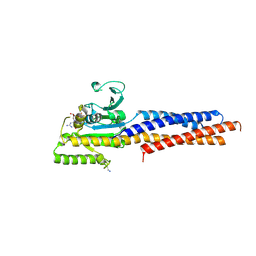 | | the structure of mini-MFN1 K88A in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Mitofusin-1 | | Authors: | Yan, L, Yu, C, Ming, Z, Lou, Z, Rao, Z, Hu, J. | | Deposit date: | 2016-07-24 | | Release date: | 2016-11-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | BDLP-like folding of Mitofusin 1
To Be Published
|
|
5VKA
 
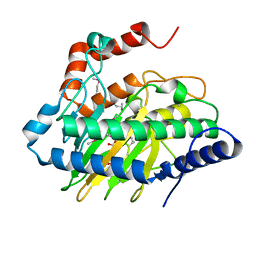 | | Ethylene forming enzyme in complex with manganese, 2-oxoglutarate and N-omega-hydroxy-L-arginine | | Descriptor: | 2-OXOGLUTARIC ACID, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, MANGANESE (II) ION, ... | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2017-04-21 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.169 Å) | | Cite: | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
5V31
 
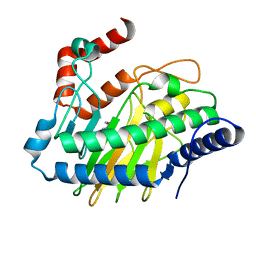 | | Ethylene forming enzyme in complex with manganese and L-arginine | | Descriptor: | 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ARGININE, MANGANESE (II) ION | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
4FO9
 
 | | Crystal structure of the E3 SUMO Ligase PIAS2 | | Descriptor: | E3 SUMO-protein ligase PIAS2, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Dong, A, Hu, J, Dobrovetsky, E, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-20 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structure of the E3 SUMO Ligase PIAS2
to be published
|
|
5V2T
 
 | | Ethylene forming enzyme in complex with manganese and tartrate | | Descriptor: | 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, L(+)-TARTARIC ACID, MANGANESE (II) ION | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2017-03-06 | | Release date: | 2017-08-16 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.227 Å) | | Cite: | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
5VKB
 
 | | Ethylene forming enzyme in complex with manganese, 2-oxoglutarate and argininamide | | Descriptor: | 2-OXOGLUTARIC ACID, 2-oxoglutarate-dependent ethylene/succinate-forming enzyme, ARGININEAMIDE, ... | | Authors: | Fellner, M, Martinez, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2017-04-21 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.139 Å) | | Cite: | Structures and Mechanisms of the Non-Heme Fe(II)- and 2-Oxoglutarate-Dependent Ethylene-Forming Enzyme: Substrate Binding Creates a Twist.
J. Am. Chem. Soc., 139, 2017
|
|
2K4K
 
 | | Solution structure of GSP13 from Bacillus subtilis | | Descriptor: | General stress protein 13 | | Authors: | Yu, W, Yu, B, Hu, J, Jin, C, Xia, B. | | Deposit date: | 2008-06-13 | | Release date: | 2009-05-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of GSP13 from Bacillus subtilis exhibits an S1 domain related to cold shock proteins.
J.Biomol.Nmr, 43, 2009
|
|
2LTU
 
 | |
7E2R
 
 | | The ligand-free structure of Arabidopsis thaliana GUN4 | | Descriptor: | Tetrapyrrole-binding protein, chloroplastic | | Authors: | Liu, L, Hu, J. | | Deposit date: | 2021-02-07 | | Release date: | 2021-08-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis of bilin binding by the chlorophyll biosynthesis regulator GUN4.
Protein Sci., 30, 2021
|
|
6JTG
 
 | |
5YDT
 
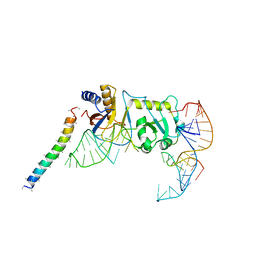 | | Remodeled Utp30 in 90S pre-ribosome (Mtr4-depleted, Enp1-TAP) | | Descriptor: | 5' ETS RNA, Ribosome biogenesis protein UTP30, Saccharomyces cerevisiae strain ALI 308 18S ribosomal RNA gene, ... | | Authors: | Ye, K, Zhu, X, Hu, J. | | Deposit date: | 2017-09-14 | | Release date: | 2017-11-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structure and RNA recognition of ribosome assembly factor Utp30.
RNA, 23, 2017
|
|
5YOV
 
 | | Crystal structure of BRD4-BD1 bound with hjp126 | | Descriptor: | (3~{R})-4-cyclopentyl-~{N}-(2,4-dimethylphenyl)-1,3-dimethyl-2-oxidanylidene-3~{H}-quinoxaline-6-carboxamide, Bromodomain-containing protein 4 | | Authors: | Xiong, B, Hu, J, Li, Y, Cao, D. | | Deposit date: | 2017-10-31 | | Release date: | 2018-11-07 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | Structure-based optimization of a series of selective BET inhibitors containing aniline or indoline groups.
Eur.J.Med.Chem., 150, 2018
|
|
