5BRQ
 
 | |
5BRP
 
 | |
4S05
 
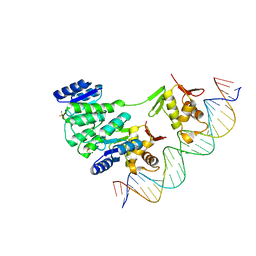 | | Crystal structure of Klebsiella pneumoniae PmrA in complex with PmrA box DNA | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DNA (26-MER), DNA-binding transcriptional regulator BasR, ... | | Authors: | Hsiao, C.D, Weng, T.H, Li, Y.C. | | Deposit date: | 2014-12-30 | | Release date: | 2015-11-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structure and dynamics of polymyxin-resistance-associated response regulator PmrA in complex with promoter DNA.
Nat Commun, 6, 2015
|
|
4S04
 
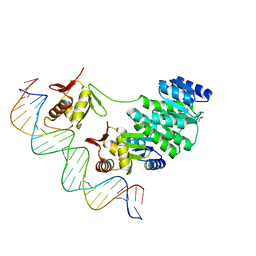 | | Crystal structure of Klebsiella pneumoniae PmrA in complex with PmrA box DNA | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, DNA (25-MER), DNA-binding transcriptional regulator BasR, ... | | Authors: | Hsiao, C.D, Weng, T.H, Li, Y.C. | | Deposit date: | 2014-12-30 | | Release date: | 2015-11-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure and dynamics of polymyxin-resistance-associated response regulator PmrA in complex with promoter DNA.
Nat Commun, 6, 2015
|
|
1E9L
 
 | |
1GGG
 
 | |
1F3V
 
 | | Crystal structure of the complex between the N-terminal domain of TRADD and the TRAF domain of TRAF2 | | Descriptor: | TUMOR NECROSIS FACTOR RECEPTOR TYPE 1 ASSOCIATED DEATH DOMAIN PROTEIN, TUMOR NECROSIS FACTOR RECEPTOR-ASSOCIATED PROTEIN | | Authors: | Park, Y.C, Ye, H, Hsia, C, Segal, D, Rich, R, Liou, H.-C, Myszka, D, Wu, H. | | Deposit date: | 2000-06-06 | | Release date: | 2000-09-06 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A novel mechanism of TRAF signaling revealed by structural and functional analyses of the TRADD-TRAF2 interaction.
Cell(Cambridge,Mass.), 101, 2000
|
|
1Q5P
 
 | | S156E/S166D variant of Bacillus lentus subtilisin | | Descriptor: | CALCIUM ION, SULFATE ION, Serine protease | | Authors: | Bott, R.R, Chan, G, Domingo, B, Ganshaw, G, Hsia, C.Y, Knapp, M, Murray, C.J. | | Deposit date: | 2003-08-08 | | Release date: | 2003-11-11 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Do enzymes change the nature of transition states? Mapping the transition state for general acid-base catalysis of a serine protease
Biochemistry, 42, 2003
|
|
4OEG
 
 | | Crystal Structure Analysis of FGF2-Disaccharide (S9I2) complex | | Descriptor: | 2-deoxy-3,6-di-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-1-O-methyl-2-O-sulfo-alpha-L-idopyranuronic acid, Fibroblast growth factor 2 | | Authors: | Li, Y.C, Hsiao, C.D. | | Deposit date: | 2014-01-13 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Interactions that influence the binding of synthetic heparan sulfate based disaccharides to fibroblast growth factor-2.
Acs Chem.Biol., 9, 2014
|
|
4UMK
 
 | | The complex of Spo0J and parS DNA in chromosomal partition system | | Descriptor: | DNA, PROBABLE CHROMOSOME-PARTITIONING PROTEIN PARB, SULFATE ION | | Authors: | Chen, B.W, Chu, C.H, Tung, J.Y, Hsu, C.E, Hsiao, C.D, Sun, Y.J. | | Deposit date: | 2014-05-19 | | Release date: | 2015-05-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.096 Å) | | Cite: | Insights into ParB spreading from the complex structure of Spo0J and parS.
Proc. Natl. Acad. Sci. U.S.A., 112, 2015
|
|
8WQ8
 
 | |
8YK9
 
 | | Structure of Saccharolobus solfataricus SegC (SSO0033) protein, ATP soak | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SegC | | Authors: | Yen, C.Y, Lin, M.G, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2024-03-04 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Unraveling the structure and function of a novel SegC protein interacting with the SegAB chromosome segregation complex in Archaea.
Nucleic Acids Res., 2024
|
|
8WQN
 
 | |
2YIK
 
 | | Catalytic domain of Clostridium thermocellum CelT | | Descriptor: | CALCIUM ION, ENDOGLUCANASE, ZINC ION | | Authors: | Tsai, J.-Y, Kesavulu, M.M, Hsiao, C.-D. | | Deposit date: | 2011-05-16 | | Release date: | 2012-02-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the Catalytic Domain of the Clostridium Thermocellum Cellulase Celt
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1C72
 
 | | TYR115, GLN165 AND TRP209 CONTRIBUTE TO THE 1,2-EPOXY-3-(P-NITROPHENOXY)PROPANE CONJUGATING ACTIVITIES OF GLUTATHIONE S-TRANSFERASE CGSTM1-1 | | Descriptor: | 1-HYDROXY-2-S-GLUTATHIONYL-3-PARA-NITROPHENOXY-PROPANE, PROTEIN (GLUTATHIONE S-TRANSFERASE) | | Authors: | Chern, M.K, Wu, T.C, Hsieh, C.H, Chou, C.C, Liu, L.F, Kuan, I.C, Yeh, Y.H, Hsiao, C.D, Tam, M.F. | | Deposit date: | 2000-02-02 | | Release date: | 2000-08-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Tyr115, gln165 and trp209 contribute to the 1, 2-epoxy-3-(p-nitrophenoxy)propane-conjugating activity of glutathione S-transferase cGSTM1-1.
J.Mol.Biol., 300, 2000
|
|
7DUV
 
 | | Structure of Sulfolobus solfataricus SegB protein | | Descriptor: | SULFATE ION, SegB | | Authors: | Yen, C.Y, Lin, M.G, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2021-01-11 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
4BM5
 
 | | Chloroplast inner membrane protein TIC110 | | Descriptor: | SIMILAR TO CHLOROPLAST INNER MEMBRANE PROTEIN TIC110 | | Authors: | Tsai, J.-Y, Chu, C.-C, Yeh, Y.-H, Chen, L.-J, Li, H.-m, Hsiao, C.-D. | | Deposit date: | 2013-05-06 | | Release date: | 2013-06-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structural Characterizations of Chloroplast Translocon Protein Tic110.
Plant J., 75, 2013
|
|
3HR9
 
 | | The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase F40I mutant | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, Beta-glucanase, ... | | Authors: | Tsai, L.C, Huang, H.C, Hsiao, C.H. | | Deposit date: | 2009-06-09 | | Release date: | 2009-07-07 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase mutant F40I
To be Published
|
|
4AWX
 
 | | Moonlighting functions of FeoC in the regulation of ferrous iron transport in Feo | | Descriptor: | FERROUS IRON TRANSPORT PROTEIN B, FERROUS IRON TRANSPORT PROTEIN C, NICKEL (II) ION, ... | | Authors: | Hung, K.-W, Tsai, J.-Y, Juan, T.-H, Hsu, Y.-L, Hsiao, C.D, Huang, T.-H. | | Deposit date: | 2012-06-06 | | Release date: | 2013-03-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of the Klebsiella Pneumoniae Nfeob/Feoc Complex and Roles of Feoc in Regulation of Fe2+ Transport by the Bacterial Feo System.
J.Bacteriol., 194, 2012
|
|
1C7R
 
 | | THE CRYSTAL STRUCTURE OF PHOSPHOGLUCOSE ISOMERASE/AUTOCRINE MOTILITY FACTOR/NEUROLEUKIN COMPLEXED WITH ITS CARBOHYDRATE PHOSPHATE INHIBITORS AND ITS SUBSTRATE RECOGNITION MECHANISM | | Descriptor: | 5-PHOSPHOARABINONIC ACID, PHOSPHOGLUCOSE ISOMERASE | | Authors: | Chou, C.-C, Meng, M, Sun, Y.-J, Hsiao, C.-D. | | Deposit date: | 2000-03-02 | | Release date: | 2000-09-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The crystal structure of phosphoglucose isomerase/autocrine motility factor/neuroleukin complexed with its carbohydrate phosphate inhibitors suggests its substrate/receptor recognition.
J.Biol.Chem., 275, 2000
|
|
1C7Q
 
 | | THE CRYSTAL STRUCTURE OF PHOSPHOGLUCOSE ISOMERASE/AUTOCRINE MOTILITY FACTOR/NEUROLEUKIN COMPLEXED WITH ITS CARBOHYDRATE PHOSPHATE INHIBITORS AND ITS SUBSTRATE RECOGNITION MECHANISM | | Descriptor: | N-BROMOACETYL-AMINOETHYL PHOSPHATE, PHOSPHOGLUCOSE ISOMERASE | | Authors: | Chou, C.-C, Meng, M, Sun, Y.-J, Hsiao, C.-D. | | Deposit date: | 2000-03-02 | | Release date: | 2000-09-13 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of phosphoglucose isomerase/autocrine motility factor/neuroleukin complexed with its carbohydrate phosphate inhibitors suggests its substrate/receptor recognition.
J.Biol.Chem., 275, 2000
|
|
7DUT
 
 | | Structure of Sulfolobus solfataricus SegA protein | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, SOJ protein (Soj) | | Authors: | Yen, C.Y, Lin, M.G, Hsiao, C.D, Sun, Y.J. | | Deposit date: | 2021-01-11 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
7DV2
 
 | | Structure of Sulfolobus solfataricus SegB-DNA complex | | Descriptor: | DNA (5'-D(P*AP*CP*GP*TP*AP*GP*AP*AP*GP*AP*GP*TP*CP*TP*AP*GP*AP*CP*TP*G)-3'), DNA (5'-D(P*CP*AP*GP*TP*CP*TP*AP*GP*AP*CP*TP*CP*TP*TP*CP*TP*AP*CP*GP*TP*A)-3'), SegB | | Authors: | Yen, C.Y, Lin, M.G, Sun, Y.J, Hsiao, C.D. | | Deposit date: | 2021-01-12 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
7DV3
 
 | | Structure of Sulfolobus solfataricus SegA-AMPPNP protein | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SOJ protein (Soj) | | Authors: | Yen, C.Y, Lin, M.G, Wu, C.T, Hsiao, C.D, Sun, Y.J. | | Deposit date: | 2021-01-12 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
7DWR
 
 | | Structure of Sulfolobus solfataricus SegA-ADP complex bound to DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA (5'-D(P*AP*GP*GP*GP*TP*GP*TP*TP*CP*CP*AP*CP*GP*TP*GP*AP*AP*AP*CP*AP*GP*GP*GP*A)-3'), DNA (5'-D(P*TP*CP*CP*CP*TP*GP*TP*TP*TP*CP*AP*CP*GP*TP*GP*GP*AP*AP*CP*AP*CP*CP*CP*T)-3'), ... | | Authors: | Yen, C.Y, Lin, M.G, Hsiao, C.D, Sun, Y.J. | | Deposit date: | 2021-01-17 | | Release date: | 2021-12-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chromosome segregation in Archaea: SegA- and SegB-DNA complex structures provide insights into segrosome assembly.
Nucleic Acids Res., 49, 2021
|
|
