8SAW
 
 | | CryoEM structure of DH270.UCA.G57R-CH848.10.17DT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.3.D0949.10.17chim.6R.SOSIP.664 gp120A, CH848.3.D0949.10.17chim.6R.SOSIP.664 gp41, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-02 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SAN
 
 | | CryoEM structure of VRC01-CH848.0836.10 | | Descriptor: | CH848.0836.10 gp120, CH848.0836.10 gp41, VCR01 variable heavy chain, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SAU
 
 | | CryoEM structure of DH270.4-CH848.10.17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.10.17 gp120, CH848.10.17 gp41, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-02 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SAY
 
 | | CryoEM structure of DH270.3-CH848.10.17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.10.17 gp120, CH848.10.17 gp41, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-02 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SB3
 
 | | CryoEM structure of DH270.2-CH848.10.17 | | Descriptor: | CH848.10.17 gp120, CH848.10.17.SOSIP gp41, DH270.2 Variable Heavy chain, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-02 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SAX
 
 | | CryoEM structure of DH270.UCA-CH848.10.17DT | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.10.17.SOSIP gp120, CH848.10.17.SOSIP gp41, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-02 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SAL
 
 | | CryoEM structure of VRC01-CH848.0358.80 | | Descriptor: | CH0848.3.D0358.80.06CHIM.DS.6R.SOSIP gp120, CH0848.3.D0358.80.06CHIM.DS.6R.SOSIP gp41, VCR01 variable heavy chain, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (4.9 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SAT
 
 | | CryoEM structure of VRC01-CH848.10.17 | | Descriptor: | CH848.10.17 gp120, CH848.10.17 gp41, VRC01-variable heavy chain, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-02 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SAZ
 
 | | CryoEM structure of DH270.I5.6-CH848.10.17 | | Descriptor: | CH848.10.17.SOSIP gp41, DH270.I5.6 variable heavy chain, DH270.I5.6 variable light chain, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-02 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SB4
 
 | | CryoEM structure of DH270.1-CH848.10.17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.10.17 gp120, CH848.10.17 gp41, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-02 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SAS
 
 | | CryoEM structure of DH270.5-CH848.10.17 | | Descriptor: | CH848.10.17 gp120, CH848.10.17 gp41, DH270.5 variable heavy chian, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SAQ
 
 | | CryoEM structure of DH270.6-CH848.0526.25 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.0526.25 gp120, CH848.0526.25 gp41, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-01 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SB5
 
 | | CryoEM structure of DH270.I1.6-CH848.10.17 | | Descriptor: | CH848.10.17.SOSIP gp120, CH848.10.17.SOSIP gp41, DH270.I1.6 variable heavy chain, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-02 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SB0
 
 | | CryoEM structure of DH270.I4.6-CH848.10.17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.10.17.SOSIP gp120, CH848.10.17.SOSIP gp41, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-02 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
8SB1
 
 | | CryoEM structure of DH270.I3-CH848.10.17 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CH848.10.17 gp120, CH848.10.17.SOSIP gp41, ... | | Authors: | Henderson, R, Zhou, Y, Stalls, V, Bartesaghi, B, Acharya, P. | | Deposit date: | 2023-04-02 | | Release date: | 2023-04-19 | | Last modified: | 2023-05-31 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural basis for breadth development in the HIV-1 V3-glycan targeting DH270 antibody clonal lineage.
Nat Commun, 14, 2023
|
|
6BOG
 
 | | Crystal structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription | | Descriptor: | RNA polymerase-associated protein RapA, SULFATE ION | | Authors: | Shaw, G.X, Gan, J, Zhou, Y.N, Zhang, R, Joachimiak, A, Jin, D.J, Ji, X. | | Deposit date: | 2017-11-20 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.205 Å) | | Cite: | Structure of RapA, a Swi2/Snf2 protein that recycles RNA polymerase during transcription.
Structure, 16, 2008
|
|
7CBL
 
 | | Cryo-EM structure of the flagellar LP ring from Salmonella | | Descriptor: | Flagellar L-ring protein, Flagellar P-ring protein, OCTANOIC ACID (CAPRYLIC ACID) | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-06-12 | | Release date: | 2021-04-28 | | Last modified: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CG7
 
 | | Cryo-EM structure of the flagellar MS ring with C34 symmetry from Salmonella | | Descriptor: | Flagellar M-ring protein | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-06-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.61 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CGO
 
 | | Cryo-EM structure of the flagellar motor-hook complex from Salmonella | | Descriptor: | Flagellar L-ring protein, Flagellar M-ring protein, Flagellar MS ring L1, ... | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-07-01 | | Release date: | 2021-04-28 | | Last modified: | 2021-05-26 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7DHI
 
 | | Cryo-EM structure of the partial agonist salbutamol-bound beta2 adrenergic receptor-Gs protein complex. | | Descriptor: | Beta-2 adrenergic receptor, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Yang, F, Ling, S.L, Zhou, Y.X, Zhang, Y.N, Lv, P, Liu, S.L, Fang, W, Sun, W.J, Hu, L.Y.A. | | Deposit date: | 2020-11-15 | | Release date: | 2020-12-16 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Different Conformational Responses of the beta2-Adrenergic Receptor-Gs Complex upon Binding of the Partial Agonist Salbutamol or the Full Agonist Isoprenaline
Natl Sci Rev, 2020
|
|
5KLV
 
 | | Structure of bos taurus cytochrome bc1 with fenamidone inhibited | | Descriptor: | (2R)-3-{[(S)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(tetradecanoyloxy)propyl octadecanoate, (5S)-5-methyl-2-(methylsulfanyl)-5-phenyl-3-(phenylamino)-3,5-dihydro-4H-imidazol-4-one, 1,2-DIHEXANOYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ... | | Authors: | Xia, D, Esser, L, Zhou, F, Zhou, Y, Xiao, Y, Tang, W.K, Yu, C.A, Qin, Z. | | Deposit date: | 2016-06-25 | | Release date: | 2016-10-12 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.652 Å) | | Cite: | Hydrogen Bonding to the Substrate Is Not Required for Rieske Iron-Sulfur Protein Docking to the Quinol Oxidation Site of Complex III.
J.Biol.Chem., 291, 2016
|
|
7CGB
 
 | | Cryo-EM structure of the flagellar hook from Salmonella | | Descriptor: | Flagellar hook protein FlgE | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-07-01 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CG4
 
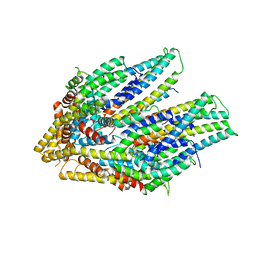 | | Cryo-EM structure of the flagellar export apparatus with FliE from Salmonella | | Descriptor: | Flagellar biosynthetic protein FliP, Flagellar hook-basal body complex protein FliE | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-06-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CBM
 
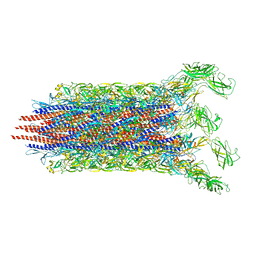 | | Cryo-EM structure of the flagellar distal rod with partial hook from Salmonella | | Descriptor: | Flagellar basal-body rod protein FlgF, Flagellar basal-body rod protein FlgG, Flagellar hook protein FlgE | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-06-12 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
7CG0
 
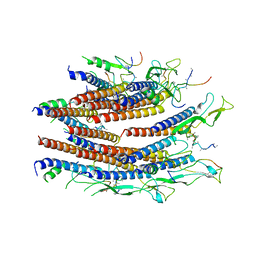 | | Cryo-EM structure of the flagellar proximal rod with FliF peptides from Salmonella | | Descriptor: | Flagellar MS ring L1, Flagellar MS ring L2, Flagellar basal body rod protein FlgB, ... | | Authors: | Tan, J.X, Chang, S.H, Wang, X.F, Xu, C.H, Zhou, Y, Zhang, X, Zhu, Y.Q. | | Deposit date: | 2020-06-30 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of assembly and torque transmission of the bacterial flagellar motor.
Cell, 184, 2021
|
|
