3FLR
 
 | |
3FLP
 
 | |
3FLT
 
 | |
6ISL
 
 | | SnoaL-like cyclase XimE with its product xiamenmycin B | | Descriptor: | (2R,3S)-2-methyl-2-(4-methylpent-3-enyl)-3-oxidanyl-3,4-dihydrochromene-6-carboxylic acid, XimE, SnoaL-like domain protein | | Authors: | He, B, Zhou, T, Bu, X, Weng, J, Xu, J, Lin, S, Zheng, J, Zhao, Y, Xu, M. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Enzymatic Pyran Formation Involved in Xiamenmycin Biosynthesis
Acs Catalysis, 9, 2019
|
|
6ISK
 
 | | SnoaL-like cyclase XimE | | Descriptor: | 1,2-ETHANEDIOL, MALONIC ACID, XimE, ... | | Authors: | He, B, Zhou, T, Bu, X, Weng, J, Xu, J, Lin, S, Zheng, J, Zhao, Y, Xu, M. | | Deposit date: | 2018-11-16 | | Release date: | 2019-11-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Enzymatic Pyran Formation Involved in Xiamenmycin Biosynthesis
Acs Catalysis, 9, 2019
|
|
8GBS
 
 | | Integrative model of the native Ana GV shell | | Descriptor: | Gas vesicle protein C, Gas vesicle structural protein | | Authors: | Dutka, P, Metskas, L.A, Hurt, R.C, Salahshoor, H, Wang, T.U, Malounda, D, Lu, G, Chou, T.F, Shapiro, M.G, Jensen, J.J. | | Deposit date: | 2023-02-28 | | Release date: | 2023-04-19 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | Structure of Anabaena flos-aquae gas vesicles revealed by cryo-ET.
Structure, 31, 2023
|
|
4NN1
 
 | |
5U9O
 
 | |
5U9L
 
 | |
3IKP
 
 | |
3IKN
 
 | |
3IKR
 
 | |
4E52
 
 | |
3IKQ
 
 | |
5D9R
 
 | | Crystal structure of a conserved domain in the intermembrane space region of the plastid division protein ARC6 | | Descriptor: | Protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic | | Authors: | Radhakrishnan, A, Kumar, N, Su, C.-C, Chou, T.-H, Yu, E. | | Deposit date: | 2015-08-18 | | Release date: | 2015-11-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.052 Å) | | Cite: | Crystal structure of a conserved domain in the intermembrane space region of the plastid division protein ARC6.
Protein Sci., 25, 2016
|
|
1GNH
 
 | | HUMAN C-REACTIVE PROTEIN | | Descriptor: | C-REACTIVE PROTEIN, CALCIUM ION | | Authors: | Shrive, A.K, Cheetham, G.M.T, Holden, D, Myles, D.A, Turnell, W.G, Volanakis, J.E, Pepys, M.B, Bloomer, A.C, Greenhough, T.J. | | Deposit date: | 1996-03-01 | | Release date: | 1997-01-27 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three dimensional structure of human C-reactive protein.
Nat.Struct.Biol., 3, 1996
|
|
4D4E
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | Descriptor: | ARMADILLO REPEAT PROTEIN ARM00016, GLYCEROL | | Authors: | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | Deposit date: | 2014-10-28 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
4D49
 
 | | Crystal structure of computationally designed armadillo repeat proteins for modular peptide recognition. | | Descriptor: | ARGININE, ARMADILLO REPEAT PROTEIN ARM00027, POLY ARG DECAPEPTIDE | | Authors: | Reichen, C, Forzani, C, Zhou, T, Parmeggiani, F, Fleishman, S.J, Mittl, P.R.E, Madhurantakam, C, Honegger, A, Ewald, C, Zerbe, O, Baker, D, Caflisch, A, Pluckthun, A. | | Deposit date: | 2014-10-27 | | Release date: | 2016-01-13 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Computationally Designed Armadillo Repeat Proteins for Modular Peptide Recognition.
J.Mol.Biol., 428, 2016
|
|
1PS5
 
 | | STRUCTURE OF THE MONOCLINIC C2 FORM OF HEN EGG-WHITE LYSOZYME AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | Lysozyme C, SULFATE ION | | Authors: | Majeed, S, Ofek, G, Belachew, A, Huang, C, Zhou, T, Kwong, P.D. | | Deposit date: | 2003-06-20 | | Release date: | 2003-09-09 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Enhancing Protein Crystallization through Precipitant Synergy
Structure, 11, 2003
|
|
1PWB
 
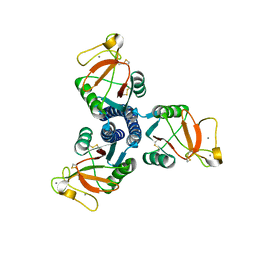 | | High resolution crystal structure of an active recombinant fragment of human lung surfactant protein D with maltose | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein D, alpha-D-glucopyranose, ... | | Authors: | Shrive, A.K, Tharia, H.A, Strong, P, Kishore, U, Burns, I, Rizkallah, P.J, Reid, K.B, Greenhough, T.J. | | Deposit date: | 2003-07-01 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | High-resolution structural insights into ligand binding and immune cell recognition by human lung surfactant protein D
J.Mol.Biol., 331, 2003
|
|
1PW9
 
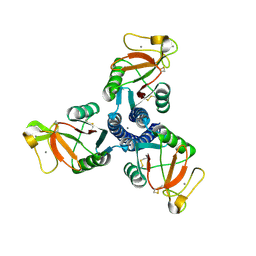 | | High resolution crystal structure of an active recombinant fragment of human lung surfactant protein D | | Descriptor: | CALCIUM ION, Pulmonary surfactant-associated protein D | | Authors: | Shrive, A.K, Tharia, H.A, Strong, P, Kishore, U, Burns, I, Rizkallah, P.J, Reid, K.B, Greenhough, T.J. | | Deposit date: | 2003-07-01 | | Release date: | 2003-08-05 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High resolution structural insights into ligand binding and immune cell recognition by human lung surfactant protein D
J.Mol.Biol., 331, 2003
|
|
1H74
 
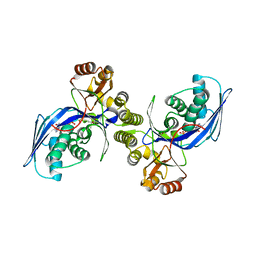 | | CRYSTAL STRUCTURE OF HOMOSERINE KINASE COMPLEXED WITH ILE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, HOMOSERINE KINASE, ISOLEUCINE, ... | | Authors: | Krishna, S.S, Zhou, T, Daugherty, M, Osterman, A.L, Zhang, H. | | Deposit date: | 2001-07-02 | | Release date: | 2001-11-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Basis for the Catalysis and Substrate Specificity of Homoserine Kinase
Biochemistry, 40, 2001
|
|
1K4M
 
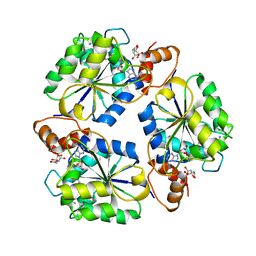 | | Crystal structure of E.coli nicotinic acid mononucleotide adenylyltransferase complexed to deamido-NAD | | Descriptor: | CITRIC ACID, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, NaMN adenylyltransferase | | Authors: | Zhang, H, Zhou, T, Kurnasov, O, Cheek, S, Grishin, N.V, Osterman, A. | | Deposit date: | 2001-10-08 | | Release date: | 2002-10-08 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structures of E. coli nicotinate mononucleotide adenylyltransferase and its complex with deamido-NAD.
Structure, 10, 2002
|
|
1K4K
 
 | | Crystal structure of E. coli Nicotinic acid mononucleotide adenylyltransferase | | Descriptor: | Nicotinic acid mononucleotide adenylyltransferase, XENON | | Authors: | Zhang, H, Zhou, T, Kurnasov, O, Cheek, S, Grishin, N.V, Osterman, A.L. | | Deposit date: | 2001-10-08 | | Release date: | 2002-10-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structures of E. coli nicotinate mononucleotide adenylyltransferase and its complex with deamido-NAD.
Structure, 10, 2002
|
|
2ZED
 
 | | Crystal structure of the human glutaminyl cyclase mutant S160A at 1.7 angstrom resolution | | Descriptor: | Glutaminyl-peptide cyclotransferase, SULFATE ION, ZINC ION | | Authors: | Huang, K.F, Wang, Y.R, Chang, E.C, Chou, T.L, Wang, A.H. | | Deposit date: | 2007-12-12 | | Release date: | 2008-04-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A conserved hydrogen-bond network in the catalytic centre of animal glutaminyl cyclases is critical for catalysis.
Biochem.J., 411, 2008
|
|
