8ES1
 
 | |
8ES0
 
 | |
4WH4
 
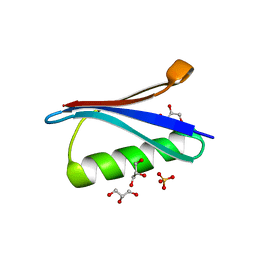 | | Protein GB1 Quadruple Mutant I6H/N8H/K28H/Q32H | | Descriptor: | GLYCEROL, Immunoglobulin G-binding protein G, SULFATE ION | | Authors: | Cunningham, T.C, Horne, W.S, Saxena, S. | | Deposit date: | 2014-09-19 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The double-histidine Cu2+-binding motif: a highly rigid, site-specific spin probe for electron spin resonance distance measurements.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
2YJ1
 
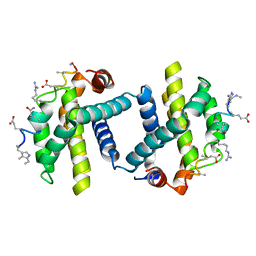 | | Puma BH3 foldamer in complex with Bcl-xL | | Descriptor: | ALPHA-BETA-PUMA BH3 FOLDAMER, BCL-2-LIKE PROTEIN 1 | | Authors: | Lee, E.F, Smith, B.J, Horne, W.S, Mayer, K.N, Evangelista, M, Colman, P.M, Gellman, S.H, Fairlie, W.D. | | Deposit date: | 2011-05-18 | | Release date: | 2011-10-12 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Structural Basis of Bcl-Xl Recognition by a Bh3-Mimetic Alpha-Beta-Peptide Generated Via Sequence-Based Design
Chembiochem, 12, 2011
|
|
6UCO
 
 | |
6UCP
 
 | |
7TIO
 
 | |
7TIR
 
 | |
7TIP
 
 | |
7TIS
 
 | |
7TIQ
 
 | |
7TV5
 
 | | Disulfide-rich venom peptide lasiocepsin: P20A mutant | | Descriptor: | Lasiocepsin | | Authors: | Cabalteja, C.C, Harmon, T.H, Rao, S.R, Horne, W.S. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Proteomimetic Analogues of Lasiocepsin, a Disulfide-Rich Antimicrobial Peptide with a Compact Tertiary Fold.
Acs Chem.Biol., 17, 2022
|
|
7TV8
 
 | | Heterogeneous-backbone proteomimetic analogue of the disulfide-rich venom peptide lasiocepsin: D-Ala modified loop | | Descriptor: | Lasiocepsin heterogeneous-backbone proteomimetic analogue | | Authors: | Cabalteja, C.C, Harmon, T.H, Rao, S.R, Horne, W.S. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Proteomimetic Analogues of Lasiocepsin, a Disulfide-Rich Antimicrobial Peptide with a Compact Tertiary Fold.
Acs Chem.Biol., 17, 2022
|
|
7TV7
 
 | | Heterogeneous-backbone proteomimetic analogue of the disulfide-rich venom peptide lasiocepsin: beta-3-Lys modified loop | | Descriptor: | Lasiocepsin heterogeneous-backbone proteomimetic analogue | | Authors: | Cabalteja, C.C, Harmon, T.H, Rao, S.R, Horne, W.S. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Proteomimetic Analogues of Lasiocepsin, a Disulfide-Rich Antimicrobial Peptide with a Compact Tertiary Fold.
Acs Chem.Biol., 17, 2022
|
|
7TV6
 
 | | Heterogeneous-backbone proteomimetic analogue of the disulfide-rich venom peptide lasiocepsin: native loop | | Descriptor: | Lasiocepsin heterogeneous-backbone proteomimetic analogue | | Authors: | Cabalteja, C.C, Harmon, T.H, Rao, S.R, Horne, W.S. | | Deposit date: | 2022-02-04 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-15 | | Method: | SOLUTION NMR | | Cite: | Heterogeneous-Backbone Proteomimetic Analogues of Lasiocepsin, a Disulfide-Rich Antimicrobial Peptide with a Compact Tertiary Fold.
Acs Chem.Biol., 17, 2022
|
|
7URJ
 
 | |
8UMS
 
 | |
8UMB
 
 | |
8UMA
 
 | |
8UM7
 
 | |
8UM9
 
 | |
9BB1
 
 | |
9BB7
 
 | |
9BB5
 
 | |
9BB4
 
 | |
