1NS4
 
 | |
1NSX
 
 | |
1NSV
 
 | |
1NS2
 
 | |
1NSR
 
 | |
1NSS
 
 | |
1NSM
 
 | |
1NSZ
 
 | |
1QOA
 
 | | FERREDOXIN MUTATION C49S | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN | | Authors: | Holden, H.M, Benning, M.M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Iron-sulfur cluster cysteine-to-serine mutants of Anabaena -2Fe-2S- ferredoxin exhibit unexpected redox properties and are competent in electron transfer to ferredoxin:NADP+ reductase.
Biochemistry, 36, 1997
|
|
1QOB
 
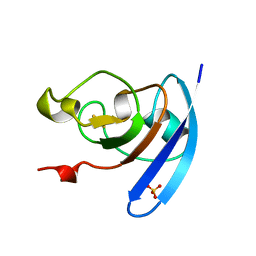 | | FERREDOXIN MUTATION D62K | | Descriptor: | FE2/S2 (INORGANIC) CLUSTER, FERREDOXIN, SULFATE ION | | Authors: | Holden, H.M, Benning, M.M. | | Deposit date: | 1997-08-14 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-function relationships in Anabaena ferredoxin: correlations between X-ray crystal structures, reduction potentials, and rate constants of electron transfer to ferredoxin:NADP+ reductase for site-specific ferredoxin mutants.
Biochemistry, 36, 1997
|
|
1HZY
 
 | | HIGH RESOLUTION STRUCTURE OF THE ZINC-CONTAINING PHOSPHOTRIESTERASE FROM PSEUDOMONAS DIMINUTA | | Descriptor: | 1,2-ETHANEDIOL, 2-PHENYL-ETHANOL, FORMIC ACID, ... | | Authors: | Holden, H.M, Benning, M.M, Raushel, F.M, Shim, H. | | Deposit date: | 2001-01-26 | | Release date: | 2001-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High resolution X-ray structures of different metal-substituted forms of phosphotriesterase from Pseudomonas diminuta.
Biochemistry, 40, 2001
|
|
1I0B
 
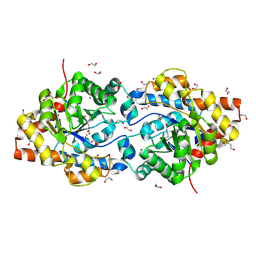 | | HIGH RESOLUTION STRUCTURE OF THE MANGANESE-CONTAINING PHOSPHOTRIESTERASE FROM PSEUDOMONAS DIMINUTA | | Descriptor: | 1,2-ETHANEDIOL, 2-PHENYL-ETHANOL, FORMIC ACID, ... | | Authors: | Holden, H.M, Benning, M.M, Raushel, F.M, Shim, H. | | Deposit date: | 2001-01-29 | | Release date: | 2001-04-04 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High resolution X-ray structures of different metal-substituted forms of phosphotriesterase from Pseudomonas diminuta.
Biochemistry, 40, 2001
|
|
1I0D
 
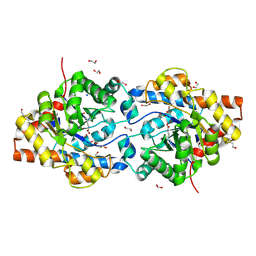 | | HIGH RESOLUTION STRUCTURE OF THE ZINC/CADMIUM-CONTAINING PHOSPHOTRIESTERASE FROM PSEUDOMONAS DIMINUTA | | Descriptor: | 1,2-ETHANEDIOL, 2-PHENYL-ETHANOL, CADMIUM ION, ... | | Authors: | Holden, H.M, Benning, M.M, Raushel, F.M, Shim, H. | | Deposit date: | 2001-01-29 | | Release date: | 2001-04-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | High resolution X-ray structures of different metal-substituted forms of phosphotriesterase from Pseudomonas diminuta.
Biochemistry, 40, 2001
|
|
1KAN
 
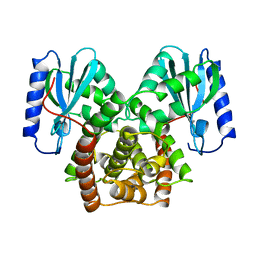 | |
1L7J
 
 | |
1L7K
 
 | |
1ISU
 
 | |
1MUS
 
 | | crystal structure of Tn5 transposase complexed with resolved outside end DNA | | Descriptor: | 1,2-ETHANEDIOL, DNA non-transferred strand, DNA transferred strand, ... | | Authors: | Holden, H.M, Thoden, J.B, Steiniger-White, M, Reznikoff, W.S, Lovell, S, Rayment, I. | | Deposit date: | 2002-09-24 | | Release date: | 2002-09-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure/function insights into Tn5 transposition.
Curr.Opin.Struct.Biol., 14, 2004
|
|
5VYS
 
 | | Crystal structure of the WbkC N-formyltransferase (C47S variant) from Brucella melitensis | | Descriptor: | 1,2-ETHANEDIOL, GUANOSINE, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Riegert, A.S, Chantigian, D.P, Thoden, J.B, Holden, H.M. | | Deposit date: | 2017-05-26 | | Release date: | 2017-07-05 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Biochemical Characterization of WbkC, an N-Formyltransferase from Brucella melitensis.
Biochemistry, 56, 2017
|
|
8EWU
 
 | | X-ray structure of the GDP-6-deoxy-4-keto-D-lyxo-heptose-4-reductase from Campylobacter jejuni HS:15 | | Descriptor: | 1,2-ETHANEDIOL, GDP-L-fucose synthase, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Xiang, D.F, Ghosh, M.K, Riegert, A.S, Raushel, F.M, Holden, H.M. | | Deposit date: | 2022-10-24 | | Release date: | 2022-11-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Bifunctional Epimerase/Reductase Enzymes Facilitate the Modulation of 6-Deoxy-Heptoses Found in the Capsular Polysaccharides of Campylobacter jejuni.
Biochemistry, 62, 2023
|
|
6TMN
 
 | | Structures of two thermolysin-inhibitor complexes that differ by a single hydrogen bond | | Descriptor: | CALCIUM ION, N-[(2R,4S)-4-hydroxy-2-(2-methylpropyl)-4-oxido-7-oxo-9-phenyl-3,8-dioxa-6-aza-4-phosphanonan-1-oyl]-L-leucine, THERMOLYSIN, ... | | Authors: | Tronrud, D.E, Holden, H.M, Matthews, B.W. | | Deposit date: | 1987-06-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of two thermolysin-inhibitor complexes that differ by a single hydrogen bond.
Science, 235, 1987
|
|
4OQE
 
 | | Crystal structure of the tylM1 N,N-dimethyltransferase in complex with SAH and TDP-Fuc3NMe | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, dTDP-3-N-methylamino-3,6-dideoxygalactose, dTDP-3-amino-3,6-dideoxy-alpha-D-glucopyranose N,N-dimethyltransferase | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2014-02-08 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Production of a novel N-monomethylated dideoxysugar.
Biochemistry, 53, 2014
|
|
4O9E
 
 | | Crystal structure of QdtA, a sugar 3,4-ketoisemerase from Thermoanaerobacterium thermosaccharolyticum in complex with TDP | | Descriptor: | (2S)-1-[3-[(2S)-2-oxidanylpropoxy]-2-[[(2S)-2-oxidanylpropoxy]methyl]-2-[[(2R)-2-oxidanylpropoxy]methyl]propoxy]propan-2-ol, QdtA, THYMIDINE-5'-DIPHOSPHATE, ... | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2014-01-02 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The molecular architecture of QdtA, a sugar 3,4-ketoisomerase from Thermoanaerobacterium thermosaccharolyticum.
Protein Sci., 23, 2014
|
|
4OQD
 
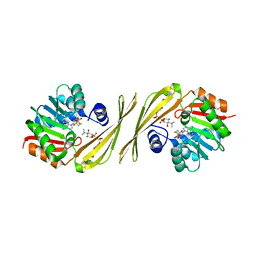 | | Crystal structure of the tylM1 N,N-dimethyltransferase in complex with SAH and TDP-Qui3NMe2 | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, TDP-3,6-dideoxy-3-N,N-dimethylglucose, dTDP-3-amino-3,6-dideoxy-alpha-D-glucopyranose N,N-dimethyltransferase | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2014-02-08 | | Release date: | 2014-02-26 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Production of a novel N-monomethylated dideoxysugar.
Biochemistry, 53, 2014
|
|
4O9G
 
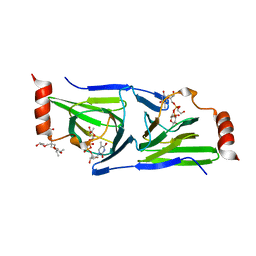 | | Crystal structure of the H51N mutant of the 3,4-ketoisomerase QdtA from Thermoanaerobacterium thermosaccharolyticum in complex with TDP-4-keto-6-deoxyglucose | | Descriptor: | (2S)-1-[3-[(2S)-2-oxidanylpropoxy]-2-[[(2S)-2-oxidanylpropoxy]methyl]-2-[[(2R)-2-oxidanylpropoxy]methyl]propoxy]propan-2-ol, 1,2-ETHANEDIOL, QdtA, ... | | Authors: | Thoden, J.B, Holden, H.M. | | Deposit date: | 2014-01-02 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The molecular architecture of QdtA, a sugar 3,4-ketoisomerase from Thermoanaerobacterium thermosaccharolyticum.
Protein Sci., 23, 2014
|
|
