7OYU
 
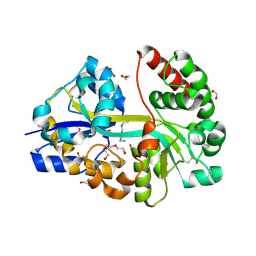 | | E.coli's putrescine receptor variant PotF/D (4JDF) with mutations E39D Y87S F88Y in complex with spermidine | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Putrescine-binding periplasmic protein PotF, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Fine-tuning spermidine binding modes in the putrescine binding protein PotF.
J.Biol.Chem., 297, 2021
|
|
7OSU
 
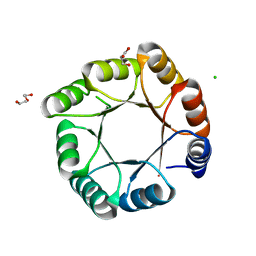 | | sTIM11noCys-SB, a de novo designed TIM barrel with a salt-bridge cluster (crystal form 1) | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Romero-Romero, S, Kordes, S, Hocker, B. | | Deposit date: | 2021-06-09 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | A newly introduced salt bridge cluster improves structural and biophysical properties of de novo TIM barrels.
Protein Sci., 31, 2022
|
|
7OSV
 
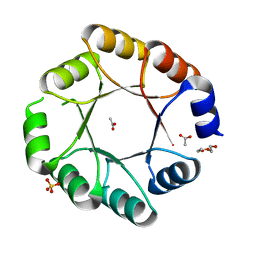 | | DeNovoTIM6-SB, a de novo designed TIM barrel with a salt-bridge cluster (crystal form 1) | | Descriptor: | ACETATE ION, DI(HYDROXYETHYL)ETHER, DeNovoTIM6-SB, ... | | Authors: | Kordes, S, Romero-Romero, S, Hocker, B. | | Deposit date: | 2021-06-09 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | A newly introduced salt bridge cluster improves structural and biophysical properties of de novo TIM barrels.
Protein Sci., 31, 2022
|
|
7P12
 
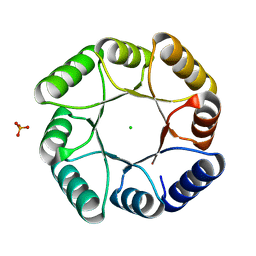 | | DeNovoTIM13-SB, a de novo designed TIM barrel with a salt-bridge cluster | | Descriptor: | CHLORIDE ION, DeNovoTIM13-SB, PHOSPHATE ION | | Authors: | Kordes, S, Romero-Romero, S, Hocker, B. | | Deposit date: | 2021-07-01 | | Release date: | 2021-12-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | A newly introduced salt bridge cluster improves structural and biophysical properties of de novo TIM barrels.
Protein Sci., 31, 2022
|
|
7OT7
 
 | |
6YED
 
 | | E.coli's Putrescine receptor PotF in its open apo state | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2020-03-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | A comprehensive binding study illustrates ligand recognition in the periplasmic binding protein PotF.
Structure, 29, 2021
|
|
6YE0
 
 | | E.coli's Putrescine receptor PotF complexed with Putrescine | | Descriptor: | (2R)-1-methoxypropan-2-amine, (2~{R})-1-[(2~{R})-1-(2-methoxyethoxy)propan-2-yl]oxypropan-2-amine, (2~{S})-1-(2-methoxyethoxy)propan-2-amine, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2020-03-23 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | A comprehensive binding study illustrates ligand recognition in the periplasmic binding protein PotF.
Structure, 29, 2021
|
|
7OYW
 
 | | E.coli's putrescine receptor variant PotF/D (4JDF) with mutations E39D F88L S247D in complex with spermidine | | Descriptor: | (2R)-1-methoxypropan-2-amine, (2~{R})-1-[(2~{R})-1-(2-methoxyethoxy)propan-2-yl]oxypropan-2-amine, (2~{S})-1-(2-methoxyethoxy)propan-2-amine, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Fine-tuning spermidine binding modes in the putrescine binding protein PotF.
J.Biol.Chem., 297, 2021
|
|
7OT8
 
 | |
6YE8
 
 | | E.coli's Putrescine receptor PotF complexed with Spermidine | | Descriptor: | (2R)-1-methoxypropan-2-amine, (2~{S})-1-(2-methoxyethoxy)propan-2-amine, CHLORIDE ION, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2020-03-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A comprehensive binding study illustrates ligand recognition in the periplasmic binding protein PotF.
Structure, 29, 2021
|
|
6YEB
 
 | | E.coli's Putrescine receptor PotF in its closed apo state | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2020-03-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | A comprehensive binding study illustrates ligand recognition in the periplasmic binding protein PotF.
Structure, 29, 2021
|
|
6YEC
 
 | | E.coli's Putrescine receptor PotF complexed with Spermine | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2020-03-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | A comprehensive binding study illustrates ligand recognition in the periplasmic binding protein PotF.
Structure, 29, 2021
|
|
2WRZ
 
 | | Crystal structure of an arabinose binding protein with designed serotonin binding site in open, ligand-free state | | Descriptor: | L-ARABINOSE-BINDING PERIPLASMIC PROTEIN | | Authors: | Schreier, B, Stumpp, C, Wiesner, S, Hocker, B. | | Deposit date: | 2009-09-03 | | Release date: | 2009-10-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Computational Design of Ligand Binding is not a Solved Problem
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6T31
 
 | | Streptavidin variants harbouring an artificial organocatalyst based cofactor | | Descriptor: | 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-(1-pyridin-4-ylpiperidin-4-yl)pentanamide, Streptavidin | | Authors: | Lechner, H, Hocker, B. | | Deposit date: | 2019-10-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | An Artificial Cofactor Catalyzing the Baylis-Hillman Reaction with Designed Streptavidin as Protein Host*.
Chembiochem, 22, 2021
|
|
7OYX
 
 | | E.coli's putrescine receptor variant PotF/D (4JDF) with mutations E39D Y87S F88Y S247D in complex with spermidine | | Descriptor: | (2~{R})-1-(2-methoxyethoxy)propan-2-amine, (2~{R})-1-[(2~{R})-1-[(2~{S})-1-[(2~{S})-1-(2-methoxyethoxy)propan-2-yl]oxypropan-2-yl]oxypropan-2-yl]oxypropan-2-amine, (2~{S})-1-(2-methoxyethoxy)propan-2-amine, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Fine-tuning spermidine binding modes in the putrescine binding protein PotF.
J.Biol.Chem., 297, 2021
|
|
6YE6
 
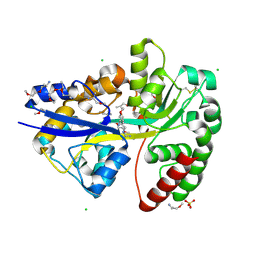 | | E.coli's Putrescine receptor PotF complexed with Agmatine | | Descriptor: | (2R)-1-methoxypropan-2-amine, (2~{R})-1-[(2~{R})-1-(2-methoxyethoxy)propan-2-yl]oxypropan-2-amine, (2~{S})-1-(2-methoxyethoxy)propan-2-amine, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2020-03-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | A comprehensive binding study illustrates ligand recognition in the periplasmic binding protein PotF.
Structure, 29, 2021
|
|
6YE7
 
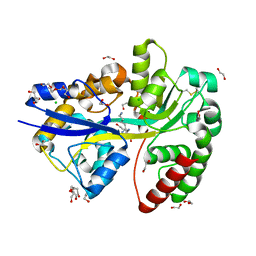 | | E.coli's Putrescine receptor PotF complexed with Cadaverine | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2020-03-24 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A comprehensive binding study illustrates ligand recognition in the periplasmic binding protein PotF.
Structure, 29, 2021
|
|
6T2Y
 
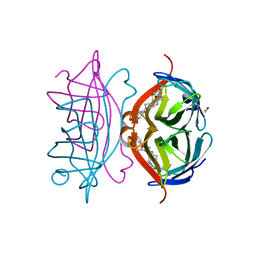 | | Streptavidin variants harbouring an artificial organocatalyst based cofactor | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-(1-pyridin-4-ylpiperidin-4-yl)pentanamide, Streptavidin | | Authors: | Lechner, H, Hocker, B. | | Deposit date: | 2019-10-10 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | An Artificial Cofactor Catalyzing the Baylis-Hillman Reaction with Designed Streptavidin as Protein Host*.
Chembiochem, 22, 2021
|
|
6T1G
 
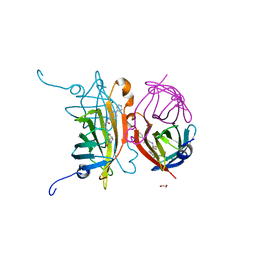 | | Streptavidin variants harbouring an artificial organocatalyst based cofactor | | Descriptor: | 1,2-ETHANEDIOL, 5-[(3~{a}~{S},4~{S},6~{a}~{R})-2-oxidanylidene-1,3,3~{a},4,6,6~{a}-hexahydrothieno[3,4-d]imidazol-4-yl]-~{N}-(1-pyridin-4-ylpiperidin-4-yl)pentanamide, Streptavidin | | Authors: | Lechner, H, Hocker, B. | | Deposit date: | 2019-10-04 | | Release date: | 2020-10-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An Artificial Cofactor Catalyzing the Baylis-Hillman Reaction with Designed Streptavidin as Protein Host*.
Chembiochem, 22, 2021
|
|
7OYV
 
 | | E.coli's putrescine receptor variant PotF/D (4JDF) with mutations E39D F88A S247D in complex with spermidine | | Descriptor: | (2R)-1-methoxypropan-2-amine, (2~{R})-1-[(2~{R})-1-[(2~{R})-1-(2-methoxyethoxy)propan-2-yl]oxypropan-2-yl]oxypropan-2-amine, (2~{S})-1-methoxypropan-2-amine, ... | | Authors: | Shanmugaratnam, S, Kroeger, P, Hocker, B. | | Deposit date: | 2021-06-25 | | Release date: | 2021-12-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Fine-tuning spermidine binding modes in the putrescine binding protein PotF.
J.Biol.Chem., 297, 2021
|
|
2XH2
 
 | |
2XGZ
 
 | |
2XH7
 
 | |
2XH0
 
 | |
2XH4
 
 | |
