5BT2
 
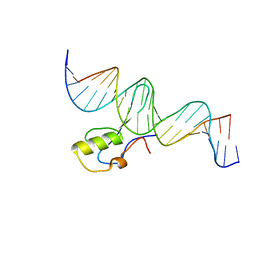 | | MeCP2 MBD domain (A140V) in complex with methylated DNA | | Descriptor: | DNA (5'-D(*AP*TP*AP*GP*AP*AP*GP*AP*AP*TP*TP*CP*(5CM)P*GP*TP*TP*CP*CP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*GP*AP*AP*(5CM)P*GP*GP*AP*AP*TP*TP*CP*TP*TP*CP*TP*A)-3'), Methyl-CpG-binding protein 2 | | Authors: | Ho, K.L, Chia, J.Y, Tan, W.S, Ng, C.L, Hu, N.J, Foo, H.L. | | Deposit date: | 2015-06-02 | | Release date: | 2016-08-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A/T Run Geometry of B-form DNA Is Independent of Bound Methyl-CpG Binding Domain, Cytosine Methylation and Flanking Sequence.
Sci Rep, 6, 2016
|
|
3C2I
 
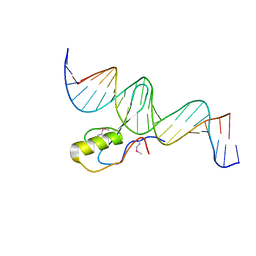 | | The Crystal Structure of Methyl-CpG Binding Domain of Human MeCP2 in Complex with a Methylated DNA Sequence from BDNF | | Descriptor: | DNA (5'-D(*DAP*DTP*DAP*DGP*DAP*DAP*DGP*DAP*DAP*DTP*DTP*DCP*(5CM)P*DGP*DTP*DTP*DCP*DCP*DAP*DG)-3'), DNA (5'-D(*DTP*DCP*DTP*DGP*DGP*DAP*DAP*(5CM)P*DGP*DGP*DAP*DAP*DTP*DTP*DCP*DTP*DTP*DCP*DTP*DA)-3'), Methyl-CpG-binding protein 2 | | Authors: | Ho, K.L, McNae, I.W, Schmiedeberg, L, Klose, R.J, Bird, A.P, Walkinshaw, M.D. | | Deposit date: | 2008-01-25 | | Release date: | 2008-05-13 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MeCP2 binding to DNA depends upon hydration at methyl-CpG
Mol.Cell, 29, 2008
|
|
5A2B
 
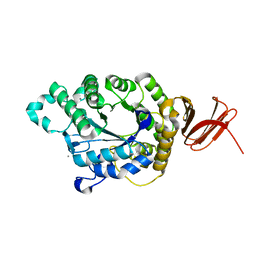 | | Crystal Structure of Anoxybacillus Alpha-amylase Provides Insights into a New Glycosyl Hydrolase Subclass | | Descriptor: | ANOXYBACILLUS ALPHA-AMYLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ng, C.L, Chai, K.P, Othman, N.F, Teh, A.H, Ho, K.L, Chan, K.G, Goh, K.M. | | Deposit date: | 2015-05-17 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Anoxybacillus Alpha-Amylase Provides Insights Into Maltose Binding of a New Glycosyl Hydrolase Subclass.
Sci.Rep., 6, 2016
|
|
5A2A
 
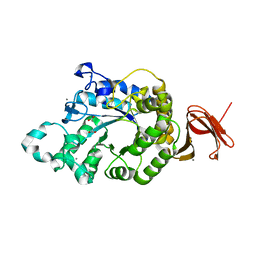 | | Crystal Structure of Anoxybacillus Alpha-amylase Provides Insights into a New Glycosyl Hydrolase Subclass | | Descriptor: | ACETATE ION, APO FORM OF ANOXYBACILLUS ALPHA-AMYLASES, CALCIUM ION | | Authors: | Ng, C.L, Chai, K.P, Othman, N.F, Teh, A.H, Ho, K.L, Chan, K.G, Goh, K.M. | | Deposit date: | 2015-05-16 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Anoxybacillus Alpha-Amylase Provides Insights Into Maltose Binding of a New Glycosyl Hydrolase Subclass.
Sci.Rep., 6, 2016
|
|
5A2C
 
 | | Crystal Structure of Anoxybacillus Alpha-amylase Provides Insights into a New Glycosyl Hydrolase Subclass | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ng, C.L, Chai, K.P, Othman, N.F, Teh, A.H, Ho, K.L, Chan, K.G, Goh, K.M. | | Deposit date: | 2015-05-17 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Anoxybacillus Alpha-Amylase Provides Insights Into Maltose Binding of a New Glycosyl Hydrolase Subclass.
Sci.Rep., 6, 2016
|
|
5YNX
 
 | | Structure of house dust mite allergen Der f 21 in PEG400 | | Descriptor: | Allergen Der f 21, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Ng, C.L, Chew, F.T, Pang, S.L, Ho, K.L, Teh, A.H, Waterman, J, Rambo, R, Mathavan, I, Beis, K, Say, Y.H. | | Deposit date: | 2017-10-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure and epitope analysis of house dust mite allergen Der f 21.
Sci Rep, 9, 2019
|
|
5YNY
 
 | | Structure of house dust mite allergen Der F 21 in PEG2KMME | | Descriptor: | Allergen Der f 21 | | Authors: | Ng, C.L, Chew, F.T, Pang, S.L, Ho, K.L, Teh, A.H, Waterman, J, Rambo, R, Mathavan, I. | | Deposit date: | 2017-10-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and epitope analysis of house dust mite allergen Der f 21.
Sci Rep, 9, 2019
|
|
5ZRT
 
 | | Crystal structure of human C1ORF123 protein | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Rahaman, S.N.A, Yusop, J.M, Mohamed-Hussein, Z.A, Wan Mohd, A, Ho, K.L, Teh, A.H, Waterman, J, Ng, C.L. | | Deposit date: | 2018-04-25 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of human C1ORF123.
Peerj, 6, 2018
|
|
2QIJ
 
 | | Hepatitis B Capsid Protein with an N-terminal extension modelled into 8.9 A data. | | Descriptor: | Core antigen | | Authors: | Tan, W.S, McNae, I.W, Ho, K.L, Walkinshaw, M.D. | | Deposit date: | 2007-07-04 | | Release date: | 2007-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (8.9 Å) | | Cite: | Crystallization and X-ray analysis of the T = 4 particle of hepatitis B capsid protein with an N-terminal extension.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
7VXR
 
 | | Crystal structure of BPSL1038 from Burkholderia pseudomallei | | Descriptor: | BPSL1038, SODIUM ION | | Authors: | Shaibullah, S, Mohd-Sharif, M, Ho, K.L, Firdaus-Raih, M, Nathan, S, Mohamed, R, Teh, A.K, Waterman, J, Ng, C.L. | | Deposit date: | 2021-11-13 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and functional analyses of Burkholderia pseudomallei BPSL1038 reveal a Cas-2/VapD nuclease sub-family.
Commun Biol, 6, 2023
|
|
7VXT
 
 | | Crystal structure of a selenomethionine-labeled BPSL1038 from Burkholderia pseudomallei | | Descriptor: | BETA-MERCAPTOETHANOL, BPSL1038, SODIUM ION | | Authors: | Shaibullah, S, Mohd-Sharif, M, Ho, K.L, Firdaus-Raih, M, Nathan, S, Mohamed, R, Teh, A.K, Waterman, J, Ng, C.L. | | Deposit date: | 2021-11-13 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and functional analyses of Burkholderia pseudomallei BPSL1038 reveal a Cas-2/VapD nuclease sub-family.
Commun Biol, 6, 2023
|
|
