6H2B
 
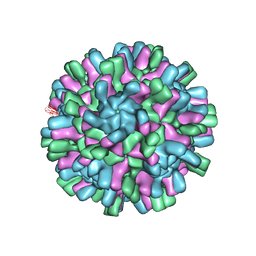 | | Structure of the Macrobrachium rosenbergii Nodavirus | | Descriptor: | CALCIUM ION, Capsid protein | | Authors: | Ho, K.H, Gabrielsen, M, Beh, P.L, Kueh, C.L, Thong, Q.X, Streetley, J, Tan, W.S, Bhella, D. | | Deposit date: | 2018-07-13 | | Release date: | 2018-10-31 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structure of the Macrobrachium rosenbergii nodavirus: A new genus within the Nodaviridae?
PLoS Biol., 16, 2018
|
|
5BT2
 
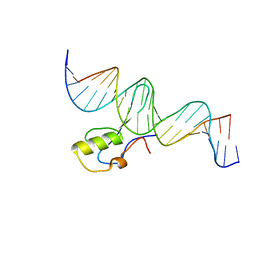 | | MeCP2 MBD domain (A140V) in complex with methylated DNA | | Descriptor: | DNA (5'-D(*AP*TP*AP*GP*AP*AP*GP*AP*AP*TP*TP*CP*(5CM)P*GP*TP*TP*CP*CP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*GP*GP*AP*AP*(5CM)P*GP*GP*AP*AP*TP*TP*CP*TP*TP*CP*TP*A)-3'), Methyl-CpG-binding protein 2 | | Authors: | Ho, K.L, Chia, J.Y, Tan, W.S, Ng, C.L, Hu, N.J, Foo, H.L. | | Deposit date: | 2015-06-02 | | Release date: | 2016-08-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A/T Run Geometry of B-form DNA Is Independent of Bound Methyl-CpG Binding Domain, Cytosine Methylation and Flanking Sequence.
Sci Rep, 6, 2016
|
|
3C2I
 
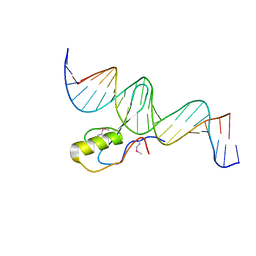 | | The Crystal Structure of Methyl-CpG Binding Domain of Human MeCP2 in Complex with a Methylated DNA Sequence from BDNF | | Descriptor: | DNA (5'-D(*DAP*DTP*DAP*DGP*DAP*DAP*DGP*DAP*DAP*DTP*DTP*DCP*(5CM)P*DGP*DTP*DTP*DCP*DCP*DAP*DG)-3'), DNA (5'-D(*DTP*DCP*DTP*DGP*DGP*DAP*DAP*(5CM)P*DGP*DGP*DAP*DAP*DTP*DTP*DCP*DTP*DTP*DCP*DTP*DA)-3'), Methyl-CpG-binding protein 2 | | Authors: | Ho, K.L, McNae, I.W, Schmiedeberg, L, Klose, R.J, Bird, A.P, Walkinshaw, M.D. | | Deposit date: | 2008-01-25 | | Release date: | 2008-05-13 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MeCP2 binding to DNA depends upon hydration at methyl-CpG
Mol.Cell, 29, 2008
|
|
6L7X
 
 | | Crystal structure of Cet1 from Trypanosoma cruzi in complex with #951 ligand | | Descriptor: | 1,3-dimethyl-7-propyl-purine-2,6-dione, SULFATE ION, mRNA_triPase domain-containing protein | | Authors: | Kuwabara, N, Ho, K. | | Deposit date: | 2019-11-03 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Crystal structures of the RNA triphosphatase fromTrypanosoma cruziprovide insights into how it recognizes the 5'-end of the RNA substrate.
J.Biol.Chem., 295, 2020
|
|
6L7W
 
 | |
6L7Y
 
 | | Crystal structure of Cet1 from Trypanosoma cruzi in complex with #466 ligand. | | Descriptor: | 3,4,6,7-tetrahydroacridine-1,8(2H,5H)-dione, SULFATE ION, mRNA_triPase domain-containing protein | | Authors: | Kuwabara, N, Ho, K. | | Deposit date: | 2019-11-03 | | Release date: | 2020-06-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structures of the RNA triphosphatase fromTrypanosoma cruziprovide insights into how it recognizes the 5'-end of the RNA substrate.
J.Biol.Chem., 295, 2020
|
|
2QIJ
 
 | | Hepatitis B Capsid Protein with an N-terminal extension modelled into 8.9 A data. | | Descriptor: | Core antigen | | Authors: | Tan, W.S, McNae, I.W, Ho, K.L, Walkinshaw, M.D. | | Deposit date: | 2007-07-04 | | Release date: | 2007-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (8.9 Å) | | Cite: | Crystallization and X-ray analysis of the T = 4 particle of hepatitis B capsid protein with an N-terminal extension.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
5YNX
 
 | | Structure of house dust mite allergen Der f 21 in PEG400 | | Descriptor: | Allergen Der f 21, BETA-MERCAPTOETHANOL, GLYCEROL, ... | | Authors: | Ng, C.L, Chew, F.T, Pang, S.L, Ho, K.L, Teh, A.H, Waterman, J, Rambo, R, Mathavan, I, Beis, K, Say, Y.H. | | Deposit date: | 2017-10-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure and epitope analysis of house dust mite allergen Der f 21.
Sci Rep, 9, 2019
|
|
5ZRT
 
 | | Crystal structure of human C1ORF123 protein | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Rahaman, S.N.A, Yusop, J.M, Mohamed-Hussein, Z.A, Wan Mohd, A, Ho, K.L, Teh, A.H, Waterman, J, Ng, C.L. | | Deposit date: | 2018-04-25 | | Release date: | 2018-08-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure and functional analysis of human C1ORF123.
Peerj, 6, 2018
|
|
5YNY
 
 | | Structure of house dust mite allergen Der F 21 in PEG2KMME | | Descriptor: | Allergen Der f 21 | | Authors: | Ng, C.L, Chew, F.T, Pang, S.L, Ho, K.L, Teh, A.H, Waterman, J, Rambo, R, Mathavan, I. | | Deposit date: | 2017-10-25 | | Release date: | 2019-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure and epitope analysis of house dust mite allergen Der f 21.
Sci Rep, 9, 2019
|
|
5A2A
 
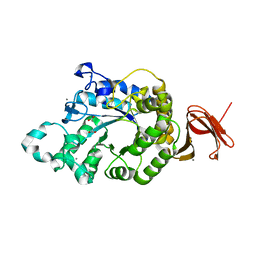 | | Crystal Structure of Anoxybacillus Alpha-amylase Provides Insights into a New Glycosyl Hydrolase Subclass | | Descriptor: | ACETATE ION, APO FORM OF ANOXYBACILLUS ALPHA-AMYLASES, CALCIUM ION | | Authors: | Ng, C.L, Chai, K.P, Othman, N.F, Teh, A.H, Ho, K.L, Chan, K.G, Goh, K.M. | | Deposit date: | 2015-05-16 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Anoxybacillus Alpha-Amylase Provides Insights Into Maltose Binding of a New Glycosyl Hydrolase Subclass.
Sci.Rep., 6, 2016
|
|
5A2B
 
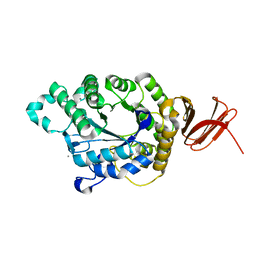 | | Crystal Structure of Anoxybacillus Alpha-amylase Provides Insights into a New Glycosyl Hydrolase Subclass | | Descriptor: | ANOXYBACILLUS ALPHA-AMYLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ng, C.L, Chai, K.P, Othman, N.F, Teh, A.H, Ho, K.L, Chan, K.G, Goh, K.M. | | Deposit date: | 2015-05-17 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Anoxybacillus Alpha-Amylase Provides Insights Into Maltose Binding of a New Glycosyl Hydrolase Subclass.
Sci.Rep., 6, 2016
|
|
5A2C
 
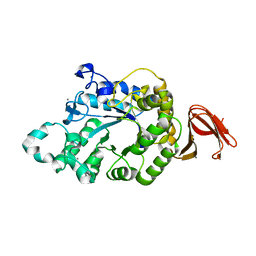 | | Crystal Structure of Anoxybacillus Alpha-amylase Provides Insights into a New Glycosyl Hydrolase Subclass | | Descriptor: | ALPHA-AMYLASE, CALCIUM ION, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Ng, C.L, Chai, K.P, Othman, N.F, Teh, A.H, Ho, K.L, Chan, K.G, Goh, K.M. | | Deposit date: | 2015-05-17 | | Release date: | 2016-03-30 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of Anoxybacillus Alpha-Amylase Provides Insights Into Maltose Binding of a New Glycosyl Hydrolase Subclass.
Sci.Rep., 6, 2016
|
|
6L7V
 
 | |
7VXT
 
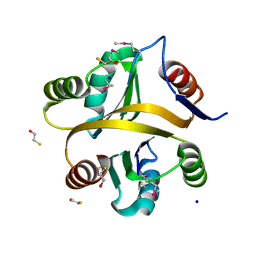 | | Crystal structure of a selenomethionine-labeled BPSL1038 from Burkholderia pseudomallei | | Descriptor: | BETA-MERCAPTOETHANOL, BPSL1038, SODIUM ION | | Authors: | Shaibullah, S, Mohd-Sharif, M, Ho, K.L, Firdaus-Raih, M, Nathan, S, Mohamed, R, Teh, A.K, Waterman, J, Ng, C.L. | | Deposit date: | 2021-11-13 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural and functional analyses of Burkholderia pseudomallei BPSL1038 reveal a Cas-2/VapD nuclease sub-family.
Commun Biol, 6, 2023
|
|
4EZ1
 
 | | Crystal structure of acetylcholine binding protein (AChBP) from Aplysia Californica in complex with alpha-conotoxin BuIA | | Descriptor: | Alpha-conotoxin BuIA, MANGANESE (II) ION, Soluble acetylcholine receptor | | Authors: | Talley, T.T, Reger, A.S, Kim, C, Sankaran, B, Ho, K, Taylor, P, McIntosh, J.M. | | Deposit date: | 2012-05-02 | | Release date: | 2013-05-08 | | Last modified: | 2013-06-19 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Pairwise interaction of alpha-conotoxin BuIA Pro6 with the beta subunit of nicotinic acetylcholine receptor
To be Published
|
|
7VXR
 
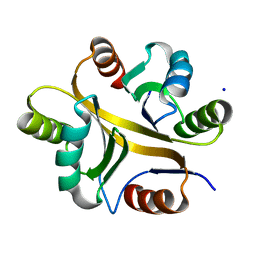 | | Crystal structure of BPSL1038 from Burkholderia pseudomallei | | Descriptor: | BPSL1038, SODIUM ION | | Authors: | Shaibullah, S, Mohd-Sharif, M, Ho, K.L, Firdaus-Raih, M, Nathan, S, Mohamed, R, Teh, A.K, Waterman, J, Ng, C.L. | | Deposit date: | 2021-11-13 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and functional analyses of Burkholderia pseudomallei BPSL1038 reveal a Cas-2/VapD nuclease sub-family.
Commun Biol, 6, 2023
|
|
2JN9
 
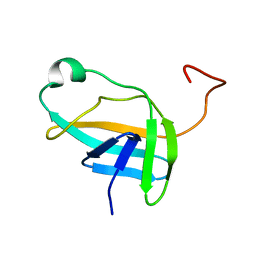 | | NMR solution structure of YkvR protein from Bacillus subtilis: NESG target SR358 | | Descriptor: | YkvR protein | | Authors: | Gurla, S.V.T, Aramini, J.M, Chi Ho, K, Cunningham, K, Ma, L.-C, Xiao, R, Baran, M.C, Acton, T.B, Liu, J, Rost, B, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-12-29 | | Release date: | 2007-05-01 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of YkvR protein from Bacillus subtilis: NESG target SR358
To be Published
|
|
4MF3
 
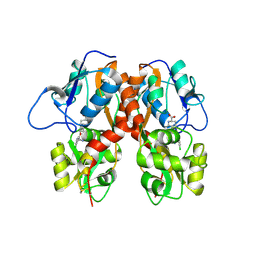 | | Crystal Structure of Human GRIK1 complexed with a 6-(tetrazolyl)aryl decahydroisoquinoline antagonist | | Descriptor: | (3S,4aS,6S,8aR)-6-[3-chloro-2-(1H-tetrazol-5-yl)phenoxy]decahydroisoquinoline-3-carboxylic acid, Glutamate receptor ionotropic, kainate 1 | | Authors: | Martinez-Perez, J.A, Iyengar, S, Shannon, H.E, Bleakman, D, Alt, A, Clawson, D.K, Arnold, B.M, Bell, M.G, Bleisch, T.J, Castano, A.M, Del Prado, M, Dominguez, E, Escribano, A.M, Filla, S.A, Ho, K.H, Hudziak, K.J, Jones, C.K, Katofiasc, M.A, Mateo, A, Mathes, B.M, Mattiuz, E.L, Ogden, A.M.L, Phebus, L.A, Simmons, R.M.A, Stack, D.R, Stratford, R.E, Winter, M.A, Wu, Z, Ornstein, P.L. | | Deposit date: | 2013-08-27 | | Release date: | 2014-05-07 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | GluK1 antagonists from 6-(tetrazolyl)phenyl decahydroisoquinoline derivatives: in vitro profile and in vivo analgesic efficacy.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
