4WHM
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with UDP | | 分子名称: | ACETATE ION, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase, ... | | 著者 | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | 登録日 | 2014-09-23 | | 公開日 | 2015-01-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.851 Å) | | 主引用文献 | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
8W6X
 
 | | Neutron structure of [NiFe]-hydrogenase from D. vulgaris Miyazaki F in its oxidized state | | 分子名称: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FE3-S4 CLUSTER, ... | | 著者 | Hiromoto, T, Tamada, T. | | 登録日 | 2023-08-30 | | 公開日 | 2023-09-13 | | 最終更新日 | 2023-11-15 | | 実験手法 | NEUTRON DIFFRACTION (1.04 Å), X-RAY DIFFRACTION | | 主引用文献 | New insights into the oxidation process from neutron and X-ray crystal structures of an O 2 -sensitive [NiFe]-hydrogenase.
Chem Sci, 14, 2023
|
|
3H65
 
 | | The Crystal Structure of C176A Mutated [Fe]-Hydrogenase (Hmd) Holoenzyme in Complex with Methylenetetrahydromethanopterin | | 分子名称: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 5'-O-[(S)-hydroxy{[2-hydroxy-3,5-dimethyl-6-(2-oxoethyl)pyridin-4-yl]oxy}phosphoryl]guanosine, 5,10-DIMETHYLENE TETRAHYDROMETHANOPTERIN, ... | | 著者 | Hiromoto, T, Warkentin, E, Shima, S, Ermler, U. | | 登録日 | 2009-04-23 | | 公開日 | 2009-09-01 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.15 Å) | | 主引用文献 | The crystal structure of an [Fe]-hydrogenase-substrate complex reveals the framework for H2 activation.
Angew.Chem.Int.Ed.Engl., 48, 2009
|
|
4REN
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with petunidin | | 分子名称: | 2-(3,4-dihydroxy-5-methoxyphenyl)-3,5,7-trihydroxychromenium, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase | | 著者 | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | 登録日 | 2014-09-23 | | 公開日 | 2015-01-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.704 Å) | | 主引用文献 | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
4REM
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with delphinidin | | 分子名称: | 3,5,7-trihydroxy-2-(3,4,5-trihydroxyphenyl)chromenium, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase | | 著者 | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | 登録日 | 2014-09-23 | | 公開日 | 2015-01-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
4REL
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase in complex with kaempferol | | 分子名称: | 3,5,7-TRIHYDROXY-2-(4-HYDROXYPHENYL)-4H-CHROMEN-4-ONE, ACETATE ION, GLYCEROL, ... | | 著者 | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | 登録日 | 2014-09-23 | | 公開日 | 2015-01-21 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.754 Å) | | 主引用文献 | Structural basis for acceptor-substrate recognition of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
Protein Sci., 24, 2015
|
|
3F46
 
 | | The Crystal Structure of C176A Mutated [Fe]-Hydrogenase (Hmd) Holoenzyme from Methanocaldococcus jannaschii | | 分子名称: | (2S,3S)-1,4-DIMERCAPTOBUTANE-2,3-DIOL, 5'-O-[(S)-hydroxy{[2-hydroxy-3,5-dimethyl-6-(2-oxoethyl)pyridin-4-yl]oxy}phosphoryl]guanosine, 5,10-methenyltetrahydromethanopterin hydrogenase, ... | | 著者 | Hiromoto, T, Vogt, S, Warkentin, E, Thauer, R.K, Shima, S, Ermler, U. | | 登録日 | 2008-10-31 | | 公開日 | 2009-02-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | The crystal structure of C176A mutated [Fe]-hydrogenase suggests an acyl-iron ligation in the active site iron complex.
Febs Lett., 583, 2009
|
|
3F47
 
 | | The Crystal Structure of [Fe]-Hydrogenase (Hmd) Holoenzyme from Methanocaldococcus jannaschii | | 分子名称: | 5'-O-[(S)-hydroxy{[2-hydroxy-3,5-dimethyl-6-(2-oxoethyl)pyridin-4-yl]oxy}phosphoryl]guanosine, 5,10-methenyltetrahydromethanopterin hydrogenase, CARBON MONOXIDE, ... | | 著者 | Hiromoto, T, Pilak, O, Warkentin, E, Thauer, R.K, Shima, S, Ermler, U. | | 登録日 | 2008-10-31 | | 公開日 | 2009-02-10 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | The crystal structure of C176A mutated [Fe]-hydrogenase suggests an acyl-iron ligation in the active site iron complex.
Febs Lett., 583, 2009
|
|
2DKH
 
 | | Crystal structure of 3-hydroxybenzoate hydroxylase from Comamonas testosteroni, in complex with the substrate | | 分子名称: | 3-HYDROXYBENZOIC ACID, 3-hydroxybenzoate hydroxylase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Hiromoto, T, Fujiwara, S, Hosokawa, K, Yamaguchi, H. | | 登録日 | 2006-04-11 | | 公開日 | 2006-10-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal structure of 3-hydroxybenzoate hydroxylase from Comamonas testosteroni has a large tunnel for substrate and oxygen access to the active site
J.Mol.Biol., 364, 2006
|
|
2DKI
 
 | | Crystal structure of 3-hydroxybenzoate hydroxylase from Comamonas testosteroni, under pressure of xenon gas (12 atm) | | 分子名称: | 3-HYDROXYBENZOATE HYDROXYLASE, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | 著者 | Hiromoto, T, Fujiwara, S, Hosokawa, K, Yamaguchi, H. | | 登録日 | 2006-04-11 | | 公開日 | 2006-10-24 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Crystal structure of 3-hydroxybenzoate hydroxylase from Comamonas testosteroni has a large tunnel for substrate and oxygen access to the active site
J.Mol.Biol., 364, 2006
|
|
3WC4
 
 | | Crystal structure of UDP-glucose: anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea | | 分子名称: | ACETATE ION, GLYCEROL, UDP-glucose:anthocyanidin 3-O-glucosyltransferase | | 著者 | Hiromoto, T, Honjo, E, Tamada, T, Kuroki, R. | | 登録日 | 2013-05-24 | | 公開日 | 2013-10-30 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Crystal structure of UDP-glucose:anthocyanidin 3-O-glucosyltransferase from Clitoria ternatea
J.SYNCHROTRON RADIAT., 20, 2013
|
|
5XPF
 
 | |
5XPE
 
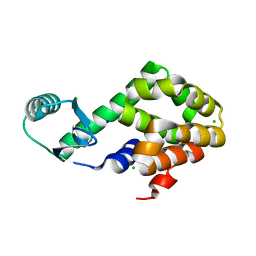 | | Neutron structure of the T26H mutant of T4 lysozyme | | 分子名称: | CHLORIDE ION, Endolysin, SODIUM ION | | 著者 | Hiromoto, T, Kuroki, R. | | 登録日 | 2017-06-01 | | 公開日 | 2017-10-04 | | 最終更新日 | 2024-04-03 | | 実験手法 | NEUTRON DIFFRACTION (1.648 Å), X-RAY DIFFRACTION | | 主引用文献 | Neutron structure of the T26H mutant of T4 phage lysozyme provides insight into the catalytic activity of the mutant enzyme and how it differs from that of wild type.
Protein Sci., 26, 2017
|
|
4DBG
 
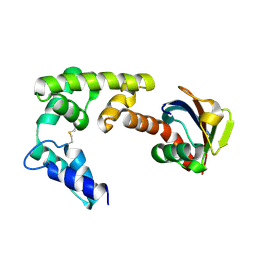 | | Crystal structure of HOIL-1L-UBL complexed with a HOIP-UBA derivative | | 分子名称: | RING finger protein 31, RanBP-type and C3HC4-type zinc finger-containing protein 1 | | 著者 | Yagi, H, Hiromoto, T, Mizushima, T, Kurimoto, E, Kato, K. | | 登録日 | 2012-01-15 | | 公開日 | 2012-04-04 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.71 Å) | | 主引用文献 | A non-canonical UBA-UBL interaction forms the linear-ubiquitin-chain assembly complex
Embo Rep., 13, 2012
|
|
3AUL
 
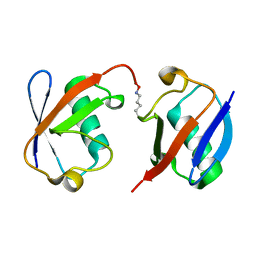 | | Crystal structure of wild-type Lys48-linked diubiquitin in an open conformation | | 分子名称: | Polyubiquitin-C | | 著者 | Hirano, T, Olivier, S, Yagi, M, Takemoto, E, Hiromoto, T, Satoh, T, Mizushima, T, Kato, K. | | 登録日 | 2011-02-09 | | 公開日 | 2011-09-07 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (2.39 Å) | | 主引用文献 | Conformational dynamics of wild-type Lys48-linked diubiquitin in solution
J.Biol.Chem., 286, 2011
|
|
7XE7
 
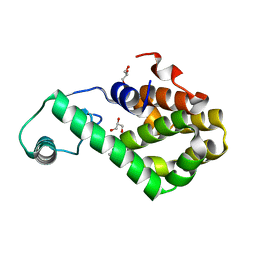 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, pH10 | | 分子名称: | Endolysin, GLYCEROL, HEXANE-1,6-DIOL | | 著者 | Tamada, T, Hiromoto, T. | | 登録日 | 2022-03-30 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-10-16 | | 実験手法 | X-RAY DIFFRACTION (1.05 Å) | | 主引用文献 | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
7XE6
 
 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, pH7 | | 分子名称: | Endolysin, GLYCEROL, HEXANE-1,6-DIOL, ... | | 著者 | Tamada, T, Hiromoto, T. | | 登録日 | 2022-03-30 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
7XEA
 
 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, DMSO 40%, and then backsoaking | | 分子名称: | CHLORIDE ION, Endolysin, GLYCEROL, ... | | 著者 | Tamada, T, Hiromoto, T. | | 登録日 | 2022-03-30 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
7XE9
 
 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, DMSO 20% | | 分子名称: | CHLORIDE ION, DIMETHYL SULFOXIDE, Endolysin, ... | | 著者 | Tamada, T, Hiromoto, T. | | 登録日 | 2022-03-30 | | 公開日 | 2023-03-22 | | 最終更新日 | 2024-10-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
7XE5
 
 | | T4 lysozyme mutant-S44C/C54T/N68C/A93C/C97A/T115C, pH4 | | 分子名称: | Endolysin, GLYCEROL, HEXANE-1,6-DIOL, ... | | 著者 | Tamada, T, Hiromoto, T. | | 登録日 | 2022-03-30 | | 公開日 | 2023-03-22 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.3 Å) | | 主引用文献 | Creation of Cross-Linked Crystals With Intermolecular Disulfide Bonds Connecting Symmetry-Related Molecules Allows Retention of Tertiary Structure in Different Solvent Conditions.
Front Mol Biosci, 9, 2022
|
|
3VR0
 
 | | Crystal structure of Pyrococcus furiosus PbaB, an archaeal proteasome activator | | 分子名称: | GOLD ION, Putative uncharacterized protein | | 著者 | Kumoi, K, Satoh, T, Hiromoto, T, Mizushima, T, Kamiya, Y, Noda, M, Uchiyama, S, Murata, K, Yagi, H, Kato, K. | | 登録日 | 2012-04-02 | | 公開日 | 2013-04-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | An archaeal homolog of proteasome assembly factor functions as a proteasome activator
Plos One, 8, 2013
|
|
3WHJ
 
 | | Crystal structure of Nas2 N-terminal domain | | 分子名称: | CADMIUM ION, Probable 26S proteasome regulatory subunit p27, SULFATE ION | | 著者 | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | 登録日 | 2013-08-26 | | 公開日 | 2014-03-26 | | 最終更新日 | 2024-05-29 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
3WHK
 
 | | Crystal structure of PAN-Rpt5C chimera | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Proteasome-activating nucleotidase, 26S protease regulatory subunit 6A | | 著者 | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | 登録日 | 2013-08-26 | | 公開日 | 2014-03-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
3WHL
 
 | | Crystal structure of Nas2 N-terminal domain complexed with PAN-Rpt5C chimera | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Probable 26S proteasome regulatory subunit p27, Proteasome-activating nucleotidase, ... | | 著者 | Satoh, T, Saeki, Y, Hiromoto, T, Wang, Y.-H, Uekusa, Y, Yagi, H, Yoshihara, H, Yagi-Utsumi, M, Mizushima, T, Tanaka, K, Kato, K. | | 登録日 | 2013-08-26 | | 公開日 | 2014-03-26 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (4 Å) | | 主引用文献 | Structural basis for proteasome formation controlled by an assembly chaperone nas2.
Structure, 22, 2014
|
|
5Y4N
 
 | | Crystal structure of aerobically purified and anaerobically crystallized D. vulgaris Miyazaki F [NiFe]-hydrogenase | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | 著者 | Nishikawa, K, Mochida, S, Hiromoto, T, Shibata, N, Higuchi, Y. | | 登録日 | 2017-08-04 | | 公開日 | 2018-08-08 | | 最終更新日 | 2024-10-23 | | 実験手法 | X-RAY DIFFRACTION (1.69 Å) | | 主引用文献 | Ni-elimination from the active site of the standard [NiFe]‐hydrogenase upon oxidation by O2.
J.Inorg.Biochem., 177, 2017
|
|
