3AYF
 
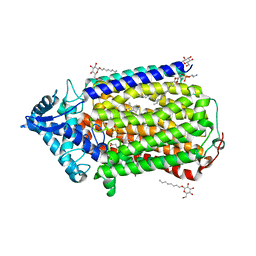 | | Crystal structure of nitric oxide reductase | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Matsumoto, Y, Tosha, T, Pisliakov, A.V, Hino, T, Sugimoti, H, Nagano, S, Sugita, Y, Shiro, Y. | | Deposit date: | 2011-05-06 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of quinol-dependent nitric oxide reductase from Geobacillus stearothermophilus
Nat.Struct.Mol.Biol., 19, 2012
|
|
3AYG
 
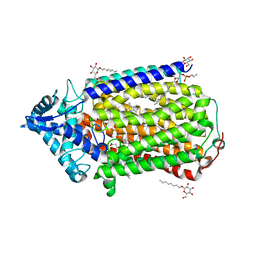 | | Crystal structure of nitric oxide reductase complex with HQNO | | Descriptor: | (1R)-2-{[(R)-(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(DODECANOYLOXY)METHYL]ETHYL (9Z)-OCTADEC-9-ENOATE, 2-HEPTYL-4-HYDROXY QUINOLINE N-OXIDE, CALCIUM ION, ... | | Authors: | Matsumoto, Y, Tosha, T, Pisliakov, A.V, Hino, T, Sugimoti, H, Nagano, S, Sugita, Y, Shiro, Y. | | Deposit date: | 2011-05-06 | | Release date: | 2012-01-25 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of quinol-dependent nitric oxide reductase from Geobacillus stearothermophilus.
Nat.Struct.Mol.Biol., 19, 2012
|
|
6KOG
 
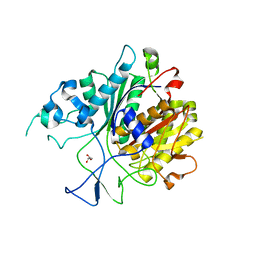 | | Ketosynthase domain in tenuazonic acid synthetase 1 (TAS1). | | Descriptor: | GLYCEROL, Hybrid PKS-NRPS synthetase TAS1, SULFATE ION | | Authors: | Yun, C.S, Nishimoto, K, Motoyama, T, Hino, T, Nagano, S, Osada, H. | | Deposit date: | 2019-08-10 | | Release date: | 2020-07-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Unique features of the ketosynthase domain in a nonribosomal peptide synthetase-polyketide synthase hybrid enzyme, tenuazonic acid synthetase 1.
J.Biol.Chem., 295, 2020
|
|
3WFE
 
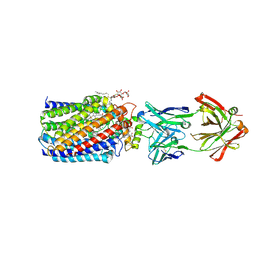 | | Reduced and cyanide-bound cytochrome c-dependent nitric oxide reductase (cNOR) from Pseudomonas aeruginosa in complex with antibody fragment | | Descriptor: | CALCIUM ION, CYANIDE ION, FE (III) ION, ... | | Authors: | Sato, N, Ishii, S, Hino, T, Sugimoto, H, Fukumori, Y, Shiro, Y, Tosha, T. | | Deposit date: | 2013-07-18 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structures of reduced and ligand-bound nitric oxide reductase provide insights into functional differences in respiratory enzymes.
Proteins, 82, 2014
|
|
3WFC
 
 | | Reduced and carbonmonoxide-bound cytochrome c-dependent nitric oxide reductase (cNOR) from Pseudomonas aeruginosa in complex with antibody fragment | | Descriptor: | CALCIUM ION, CARBON MONOXIDE, FE (III) ION, ... | | Authors: | Sato, N, Ishii, S, Hino, T, Sugimoto, H, Fukumori, Y, Shiro, Y, Tosha, T. | | Deposit date: | 2013-07-18 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of reduced and ligand-bound nitric oxide reductase provide insights into functional differences in respiratory enzymes.
Proteins, 82, 2014
|
|
3WFD
 
 | | Reduced and acetaldoxime-bound cytochrome c-dependent nitric oxide reductase (cNOR) from Pseudomonas aeruginosa in complex with antibody fragment | | Descriptor: | (1E)-N-hydroxyethanimine, CALCIUM ION, FE (III) ION, ... | | Authors: | Sato, N, Ishii, S, Hino, T, Sugimoto, H, Fukumori, Y, Shiro, Y, Tosha, T. | | Deposit date: | 2013-07-18 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of reduced and ligand-bound nitric oxide reductase provide insights into functional differences in respiratory enzymes.
Proteins, 82, 2014
|
|
3WFB
 
 | | Reduced cytochrome c-dependent nitric oxide reductase (cNOR) from Pseudomonas aeruginosa in complex with antibody fragment | | Descriptor: | CALCIUM ION, CHLORIDE ION, FE (III) ION, ... | | Authors: | Sato, N, Ishii, S, Hino, T, Sugimoto, H, Fukumori, Y, Shiro, Y, Tosha, T. | | Deposit date: | 2013-07-18 | | Release date: | 2014-05-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structures of reduced and ligand-bound nitric oxide reductase provide insights into functional differences in respiratory enzymes.
Proteins, 82, 2014
|
|
3WWX
 
 | | Crystal structure of D-stereospecific amidohydrolase from Streptomyces sp. 82F2 | | Descriptor: | OCTANE 1,8-DIAMINE, S12 family peptidase | | Authors: | Arima, J, Nagano, S, Hino, T, Shimone, K, Isoda, Y, Mori, N. | | Deposit date: | 2014-07-03 | | Release date: | 2015-07-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Crystal structure of D-stereospecific amidohydrolase from Streptomyces sp. 82F2 - insight into the structural factors for substrate specificity.
Febs J., 283, 2016
|
|
6A17
 
 | | Crystal structure of CYP90B1 in complex with brassinazole | | Descriptor: | (2R,3S)-4-(4-chlorophenyl)-2-phenyl-3-(1H-1,2,4-triazol-1-yl)butan-2-ol, CHLORIDE ION, Cytochrome P450 90B1, ... | | Authors: | Fujiyama, K, Hino, T, Kanadani, M, Mizutani, M, Nagano, S. | | Deposit date: | 2018-06-06 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Structural insights into a key step of brassinosteroid biosynthesis and its inhibition.
Nat.Plants, 5, 2019
|
|
6A15
 
 | | Structure of CYP90B1 in complex with cholesterol | | Descriptor: | CHLORIDE ION, CHOLESTEROL, Cytochrome P450 90B1, ... | | Authors: | Fujiyama, K, Hino, T, Kanadani, M, Mizutani, M, Nagano, S. | | Deposit date: | 2018-06-06 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural insights into a key step of brassinosteroid biosynthesis and its inhibition.
Nat.Plants, 5, 2019
|
|
6A18
 
 | | Crystal structure of CYP90B1 in complex with 1,6-hexandiol | | Descriptor: | CHLORIDE ION, Cytochrome P450 90B1, GLYCEROL, ... | | Authors: | Fujiyama, K, Hino, T, Kanadani, M, Mizutani, M, Nagano, S. | | Deposit date: | 2018-06-06 | | Release date: | 2019-06-12 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Structural insights into a key step of brassinosteroid biosynthesis and its inhibition.
Nat.Plants, 5, 2019
|
|
6A16
 
 | | Crystal structure of CYP90B1 in complex with uniconazole | | Descriptor: | (1E,3S)-1-(4-chlorophenyl)-4,4-dimethyl-2-(1H-1,2,4-triazol-1-yl)pent-1-en-3-ol, CHLORIDE ION, Cytochrome P450 90B1, ... | | Authors: | Fujiyama, K, Hino, T, Kanadani, M, Mizutani, M, Nagano, S. | | Deposit date: | 2018-06-06 | | Release date: | 2019-06-12 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural insights into a key step of brassinosteroid biosynthesis and its inhibition.
Nat.Plants, 5, 2019
|
|
1N5X
 
 | | Xanthine Dehydrogenase from Bovine Milk with Inhibitor TEI-6720 Bound | | Descriptor: | 2-(3-CYANO-4-ISOBUTOXY-PHENYL)-4-METHYL-5-THIAZOLE-CARBOXYLIC ACID, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Okamoto, K, Eger, B.T, Nishino, T, Kondo, S, Pai, E.F, Nishino, T. | | Deposit date: | 2002-11-07 | | Release date: | 2003-03-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | An Extremely Potent Inhibitor of Xanthine Oxidoreductase: Crystal Structure of the Enzyme-Inhibitor Complex and Mechanism of Inhibition
J.BIOL.CHEM., 278, 2003
|
|
1QQ2
 
 | | CRYSTAL STRUCTURE OF A MAMMALIAN 2-CYS PEROXIREDOXIN, HBP23. | | Descriptor: | CHLORIDE ION, THIOREDOXIN PEROXIDASE 2 | | Authors: | Hirotsu, S, Abe, Y, Okada, K, Nagahara, N, Hori, H, Nishino, T, Hakoshima, T. | | Deposit date: | 1999-06-10 | | Release date: | 1999-10-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of a multifunctional 2-Cys peroxiredoxin heme-binding protein 23 kDa/proliferation-associated gene product.
Proc.Natl.Acad.Sci.USA, 96, 1999
|
|
8Z5L
 
 | | Crystal structure of metallo-beta-lactamse, IMP-1, complexed with a quinolinone-based inhibitor | | Descriptor: | 3-[2-azanyl-5-[2-cyclohexylethyl-[3-(4-methylphenoxy)propyl]amino]phenyl]propanoic acid, Metallo-beta-lactamase type 2, ZINC ION | | Authors: | Kamo, T, Kuroda, K, Nimura, S, Guo, Y, Kondo, S, Nukaga, M, Hoshino, T. | | Deposit date: | 2024-04-18 | | Release date: | 2024-05-08 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Development of Inhibitory Compounds for Metallo-beta-lactamase through Computational Design and Crystallographic Analysis.
Biochemistry, 63, 2024
|
|
8YFY
 
 | | CRYSTAL STRUCTURE OF THE EST1 H274D MUTANT AT PH 4.2 | | Descriptor: | Carboxylesterase, octyl beta-D-glucopyranoside | | Authors: | Unno, H, Oshima, Y, Nishino, T, Nakayama, T, Kusunoki, M. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Lowering pH optimum of activity of SshEstI, a slightly alkaliphilic archaeal esterase of the hormone-sensitive lipase family.
J.Biosci.Bioeng., 138, 2024
|
|
8YFZ
 
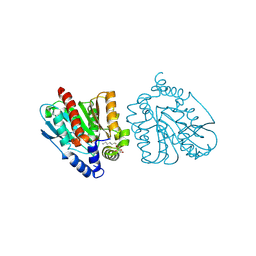 | | CRYSTAL STRUCTURE OF THE EST1 H274E MUTANT AT PH 4.2 | | Descriptor: | Carboxylesterase, octyl beta-D-glucopyranoside | | Authors: | Unno, H, Oshima, Y, Nishino, T, Nakayama, T, Kusunoki, M. | | Deposit date: | 2024-02-26 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Lowering pH optimum of activity of SshEstI, a slightly alkaliphilic archaeal esterase of the hormone-sensitive lipase family.
J.Biosci.Bioeng., 2024
|
|
1FO4
 
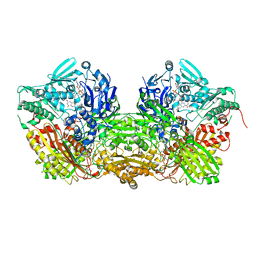 | | CRYSTAL STRUCTURE OF XANTHINE DEHYDROGENASE ISOLATED FROM BOVINE MILK | | Descriptor: | 2-HYDROXYBENZOIC ACID, CALCIUM ION, DIOXOTHIOMOLYBDENUM(VI) ION, ... | | Authors: | Enroth, C, Eger, B.T, Okamoto, K, Nishino, T, Nishino, T, Pai, E.F. | | Deposit date: | 2000-08-24 | | Release date: | 2000-10-25 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: structure-based mechanism of conversion.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1FIQ
 
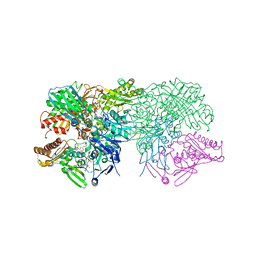 | | CRYSTAL STRUCTURE OF XANTHINE OXIDASE FROM BOVINE MILK | | Descriptor: | 2-HYDROXYBENZOIC ACID, DIOXOTHIOMOLYBDENUM(VI) ION, FE2/S2 (INORGANIC) CLUSTER, ... | | Authors: | Enroth, C, Eger, B.T, Okamoto, K, Nishino, T, Nishino, T, Pai, E.F. | | Deposit date: | 2000-08-04 | | Release date: | 2000-10-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of bovine milk xanthine dehydrogenase and xanthine oxidase: structure-based mechanism of conversion.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
6LPG
 
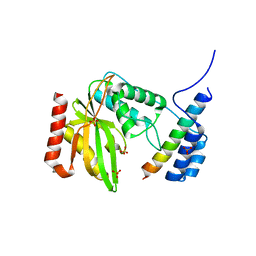 | | human VASH1-SVBP complex | | Descriptor: | SULFATE ION, Small vasohibin-binding protein, Tubulinyl-Tyr carboxypeptidase 1 | | Authors: | Ikeda, A, Nishino, T. | | Deposit date: | 2020-01-10 | | Release date: | 2020-10-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The crystal structure of the tetrameric human vasohibin-1-SVBP complex reveals a variable arm region within the structural core.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
8JYH
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with laccaic acid C | | Descriptor: | 7-[5-[(2~{S})-2-azanyl-3-oxidanyl-3-oxidanylidene-propyl]-2-oxidanyl-phenyl]-3,5,6,8-tetrakis(oxidanyl)-9,10-bis(oxidanylidene)anthracene-1,2-dicarboxylic acid, MANGANESE (II) ION, Pol protein,Pol protein,HIV-1 Reverse Transcriptase RNase H active domain, ... | | Authors: | Ito, Y, Lu, H, Kitajima, M, Ishikawa, H, Nakata, Y, Iwatani, Y, Hoshino, T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Sticklac-Derived Natural Compounds Inhibiting RNase H Activity of HIV-1 Reverse Transcriptase.
J.Nat.Prod., 86, 2023
|
|
8JYJ
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with laccaic acid A | | Descriptor: | 7-[5-(2-acetamidoethyl)-2-oxidanyl-phenyl]-3,5,6,8-tetrakis(oxidanyl)-9,10-bis(oxidanylidene)anthracene-1,2-dicarboxylic acid, MANGANESE (II) ION, Pol protein,Pol protein,HIV-1 Reverse Transcriptase RNase H active domain, ... | | Authors: | Ito, Y, Lu, H, Kitajima, M, Ishikawa, H, Nakata, Y, Iwatani, Y, Hoshino, T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Sticklac-Derived Natural Compounds Inhibiting RNase H Activity of HIV-1 Reverse Transcriptase.
J.Nat.Prod., 86, 2023
|
|
8JYI
 
 | | Crystal structure of engineered HIV-1 Reverse Transcriptase RNase H domain complexed with laccaic acid E | | Descriptor: | 7-[5-(2-azanylethyl)-2-oxidanyl-phenyl]-3,5,6,8-tetrakis(oxidanyl)-9,10-bis(oxidanylidene)anthracene-1,2-dicarboxylic acid, MANGANESE (II) ION, Pol protein,Pol protein,Ribonuclease H, ... | | Authors: | Ito, Y, Lu, H, Kitajima, M, Ishikawa, H, Nakata, Y, Iwatani, Y, Hoshino, T. | | Deposit date: | 2023-07-03 | | Release date: | 2023-08-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Sticklac-Derived Natural Compounds Inhibiting RNase H Activity of HIV-1 Reverse Transcriptase.
J.Nat.Prod., 86, 2023
|
|
5JHT
 
 | | Apo form of influenza strain H1N1 polymerase acidic subunit N-terminal region crystallized with potassium sodium tartrate | | Descriptor: | L(+)-TARTARIC ACID, MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Fudo, S, Yamamoto, N, Nukaga, M, Odagiri, T, Tashiro, M, Hoshino, T. | | Deposit date: | 2016-04-21 | | Release date: | 2016-05-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Influence of Precipitants on Molecular Arrangements and Space Groups of Protein Crystals
Cryst.Growth Des., 17, 2017
|
|
1O5P
 
 | | Solution Structure of holo-Neocarzinostatin | | Descriptor: | NEOCARZINOSTATIN-CHROMOPHORE, Neocarzinostatin | | Authors: | Takashima, H, Ishino, T, Yoshida, T, Hasuda, K, Ohkubo, T, Kobayashi, Y. | | Deposit date: | 2003-10-04 | | Release date: | 2003-10-14 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure Investigation for Releasing Mechanism of Neocarzinostatin Chromophore from the Holoprotein
J.Biol.Chem., 280, 2005
|
|
