8ROX
 
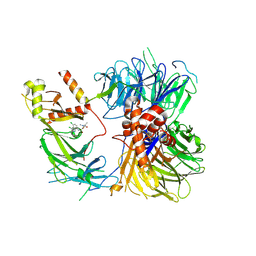 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 12 | | Descriptor: | 5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-~{N},~{N},3-trimethyl-furan-2-carboxamide;ethane, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
8ROY
 
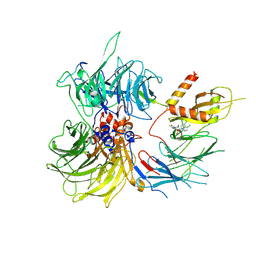 | | Structure of the human DDB1-DDA1-DCAF15 E3 ubiquitin ligase bound to compound furan 24 | | Descriptor: | 1-[5-[[3,4-bis(chloranyl)-1~{H}-indol-7-yl]sulfamoyl]-3-methyl-furan-2-yl]carbonyl-~{N}-methyl-piperidine-4-carboxamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Shilliday, F, Lucas, S.C.C, Richter, M, Michaelides, I.N, Fusani, L. | | Deposit date: | 2024-01-12 | | Release date: | 2024-04-03 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Optimization of Potent Ligands for the E3 Ligase DCAF15 and Evaluation of Their Use in Heterobifunctional Degraders.
J.Med.Chem., 67, 2024
|
|
2K7R
 
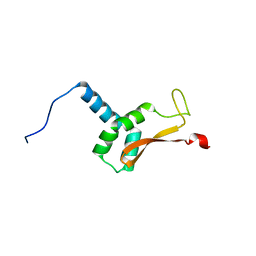 | | N-terminal domain of the Bacillus subtilis helicase-loading protein DnaI | | Descriptor: | Primosomal protein dnaI, ZINC ION | | Authors: | Loscha, K.V, Jaudzems, K, Ioannou, C, Su, X.C, Hill, F.R, Otting, G, Dixon, N.E, Liepinsh, E. | | Deposit date: | 2008-08-19 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | A novel zinc-binding fold in the helicase interaction domain of the Bacillus subtilis DnaI helicase loader
Nucleic Acids Res., 37, 2009
|
|
2YF2
 
 | |
8TC1
 
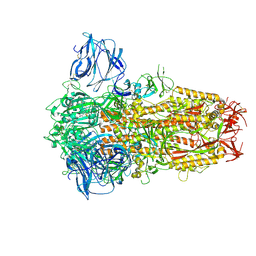 | | Cryo-EM Structure of Spike Glycoprotein from Civet Coronavirus 007 in Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Bostina, M, Hills, F.R, Eruera, A.R. | | Deposit date: | 2023-06-29 | | Release date: | 2024-05-01 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (1.92 Å) | | Cite: | Variation in structural motifs within SARS-related coronavirus spike proteins.
Plos Pathog., 20, 2024
|
|
8TC0
 
 | | Cryo-EM Structure of Spike Glycoprotein from Bat Coronavirus WIV1 in Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bostina, M, Hills, F.R, Eruera, A. | | Deposit date: | 2023-06-29 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (1.88 Å) | | Cite: | Variation in structural motifs within SARS-related coronavirus spike proteins.
Plos Pathog., 20, 2024
|
|
8TC5
 
 | | Cryo-EM Structure of Spike Glycoprotein from Civet Coronavirus SZ3 in Closed Conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bostina, M, Hills, F.R, Eruera, A. | | Deposit date: | 2023-06-29 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.11 Å) | | Cite: | Variation in structural motifs within SARS-related coronavirus spike proteins.
Plos Pathog., 20, 2024
|
|
7Z2C
 
 | | P. falciparum kinesin-8B motor domain in no nucleotide bound to tubulin dimer | | Descriptor: | Detyrosinated tubulin alpha-1B chain, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-like protein, ... | | Authors: | Liu, T, Shilliday, F, Cook, A.D, Moores, C.A. | | Deposit date: | 2022-02-26 | | Release date: | 2022-10-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Mechanochemical tuning of a kinesin motor essential for malaria parasite transmission.
Nat Commun, 13, 2022
|
|
7Z2A
 
 | | P. berghei kinesin-8B motor domain in no nucleotide state bound to tubulin dimer | | Descriptor: | Detyrosinated tubulin alpha-1B chain, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-8, ... | | Authors: | Liu, T, Shilliday, F, Cook, A.D, Moores, C.A. | | Deposit date: | 2022-02-26 | | Release date: | 2022-10-19 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mechanochemical tuning of a kinesin motor essential for malaria parasite transmission.
Nat Commun, 13, 2022
|
|
7Z2B
 
 | | P. berghei kinesin-8B motor domain in AMPPNP state bound to tubulin dimer | | Descriptor: | Detyrosinated tubulin alpha-1B chain, GUANOSINE-5'-TRIPHOSPHATE, Kinesin-8, ... | | Authors: | Liu, T, Shilliday, F, Cook, A.D, Moores, C.A. | | Deposit date: | 2022-02-26 | | Release date: | 2022-11-16 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mechanochemical tuning of a kinesin motor essential for malaria parasite transmission.
Nat Commun, 13, 2022
|
|
5TMB
 
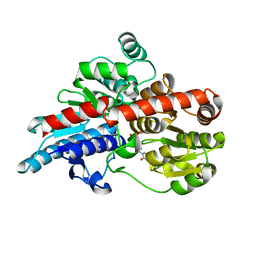 | | Crystal structure of Os79 from O. sativa in complex with UDP. | | Descriptor: | Glycosyltransferase, Os79, URIDINE-5'-DIPHOSPHATE | | Authors: | Wetterhorn, K.M, Newmister, S.A, Caniza, R.K, Busman, M, McCormick, S.P, Berthiller, F, Adam, G, Rayment, I. | | Deposit date: | 2016-10-12 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Crystal Structure of Os79 (Os04g0206600) from Oryza sativa: A UDP-glucosyltransferase Involved in the Detoxification of Deoxynivalenol.
Biochemistry, 55, 2016
|
|
2L2X
 
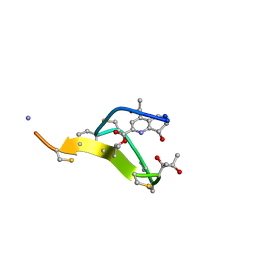 | | Thiostrepton, oxidized at CA-CB bond of residue 9 | | Descriptor: | Thiostrepton | | Authors: | Jonker, H.R.A, Baumann, S, Wolf, A, Schoof, S, Hiller, F, Schulte, K.W, Kirschner, K.N, Schwalbe, H, Arndt, H.-D. | | Deposit date: | 2010-08-27 | | Release date: | 2011-02-02 | | Last modified: | 2013-06-26 | | Method: | SOLUTION NMR | | Cite: | NMR structures of thiostrepton derivatives for characterization of the ribosomal binding site.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
2L2Z
 
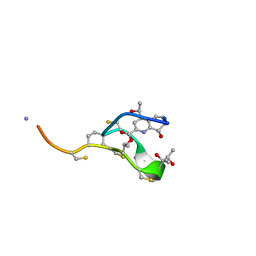 | | Thiostrepton, reduced at N-CA bond of residue 14 | | Descriptor: | Thiostrepton | | Authors: | Jonker, H.R.A, Baumann, S, Wolf, A, Schoof, S, Hiller, F, Schulte, K.W, Kirschner, K.N, Schwalbe, H, Arndt, H.-D. | | Deposit date: | 2010-08-27 | | Release date: | 2011-02-02 | | Last modified: | 2013-06-26 | | Method: | SOLUTION NMR | | Cite: | NMR structures of thiostrepton derivatives for characterization of the ribosomal binding site.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
2L2W
 
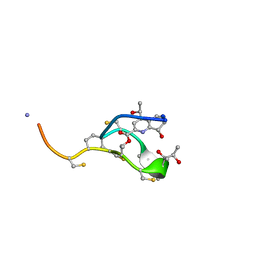 | | Thiostrepton | | Descriptor: | Thiostrepton | | Authors: | Jonker, H.R.A, Baumann, S, Wolf, A, Schoof, S, Hiller, F, Schulte, K.W, Kirschner, K.N, Schwalbe, H, Arndt, H.-D. | | Deposit date: | 2010-08-27 | | Release date: | 2011-02-02 | | Last modified: | 2013-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structures of thiostrepton derivatives for characterization of the ribosomal binding site.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
5TME
 
 | | Crystal structure of Os79 from O. sativa in complex with UDP. | | Descriptor: | Glycosyltransferase, Os79, URIDINE-5'-DIPHOSPHATE | | Authors: | Wetterhorn, K.M, Newmister, S.A, Caniza, R.K, Busman, M, McCormick, S.P, Berthiller, F, Adam, G, Rayment, I. | | Deposit date: | 2016-10-12 | | Release date: | 2016-11-02 | | Last modified: | 2022-03-16 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal Structure of Os79 (Os04g0206600) from Oryza sativa: A UDP-glucosyltransferase Involved in the Detoxification of Deoxynivalenol.
Biochemistry, 55, 2016
|
|
5TMD
 
 | | Crystal structure of Os79 from O. sativa in complex with U2F and trichothecene. | | Descriptor: | Glycosyltransferase, Os79, URIDINE-5'-DIPHOSPHATE-2-DEOXY-2-FLUORO-ALPHA-D-GLUCOSE, ... | | Authors: | Wetterhorn, K, Newmister, S.A, Caniza, R.K, Busman, M, McCormick, S.P, Berthiller, F, Adam, G, Rayment, I. | | Deposit date: | 2016-10-12 | | Release date: | 2016-11-02 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of Os79 (Os04g0206600) from Oryza sativa: A UDP-glucosyltransferase Involved in the Detoxification of Deoxynivalenol.
Biochemistry, 55, 2016
|
|
2L2Y
 
 | | Thiostrepton, epimer form of residue 9 | | Descriptor: | Thiostrepton | | Authors: | Jonker, H.R.A, Baumann, S, Wolf, A, Schoof, S, Hiller, F, Schulte, K.W, Kirschner, K.N, Schwalbe, H, Arndt, H.-D. | | Deposit date: | 2010-08-27 | | Release date: | 2011-02-02 | | Last modified: | 2013-06-26 | | Method: | SOLUTION NMR | | Cite: | NMR structures of thiostrepton derivatives for characterization of the ribosomal binding site.
Angew.Chem.Int.Ed.Engl., 50, 2011
|
|
