3PHP
 
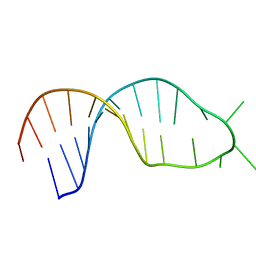 | | STRUCTURE OF THE 3' HAIRPIN OF THE TYMV PSEUDOKNOT: PREFORMATION IN RNA FOLDING | | Descriptor: | RNA (5'-R(*GP*GP*UP*UP*CP*CP*GP*AP*GP*GP*GP*UP*CP*AP*UP*CP*GP*GP*AP*AP*CP*CP*A) -3') | | Authors: | Kolk, M.H, Van Der Graaf, M, Wijmenga, S.S, Pleij, C.W.A, Heus, H.A, Hilbers, C.W. | | Deposit date: | 1998-10-13 | | Release date: | 1998-10-21 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure of the 3'-hairpin of the TYMV pseudoknot: preformation in RNA folding.
EMBO J., 17, 1998
|
|
2GVA
 
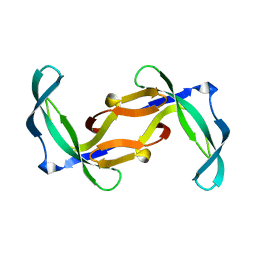 | | REFINED SOLUTION STRUCTURE OF THE TYR 41--> HIS MUTANT OF THE M13 GENE V PROTEIN. A COMPARISON WITH THE CRYSTAL STRUCTURE | | Descriptor: | GENE V PROTEIN | | Authors: | Folkers, P.J.M, Nilges, M, Folmer, R.H.A, Prompers, J.J, Konings, R.N.H, Hilbers, C.W. | | Deposit date: | 1995-07-27 | | Release date: | 1995-10-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the Tyr41-->His mutant of the M13 gene V protein. A comparison with the crystal structure.
Eur.J.Biochem., 232, 1995
|
|
2GVB
 
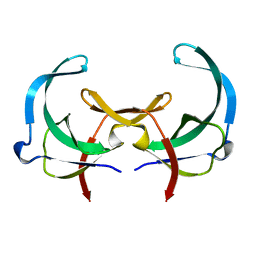 | | REFINED SOLUTION STRUCTURE OF THE TYR 41--> HIS MUTANT OF THE M13 GENE V PROTEIN. A COMPARISON WITH THE CRYSTAL STRUCTURE | | Descriptor: | GENE V PROTEIN | | Authors: | Folkers, P.J.M, Nilges, M, Folmer, R.H.A, Prompers, J.J, Konings, R.N.H, Hilbers, C.W. | | Deposit date: | 1995-07-27 | | Release date: | 1995-10-15 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Refined solution structure of the Tyr41-->His mutant of the M13 gene V protein. A comparison with the crystal structure.
Eur.J.Biochem., 232, 1995
|
|
2CPS
 
 | | SOLUTION NMR STRUCTURES OF THE MAJOR COAT PROTEIN OF FILAMENTOUS BACTERIOPHAGE M13 SOLUBILIZED IN SODIUM DODECYL SULPHATE MICELLES, 25 LOWEST ENERGY STRUCTURES | | Descriptor: | M13 MAJOR COAT PROTEIN | | Authors: | Papavoine, C.H.M, Christiaans, B.E.C, Folmer, R.H.A, Konings, R.N.H, Hilbers, C.W. | | Deposit date: | 1998-04-16 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the M13 major coat protein in detergent micelles: a basis for a model of phage assembly involving specific residues.
J.Mol.Biol., 282, 1998
|
|
2CPB
 
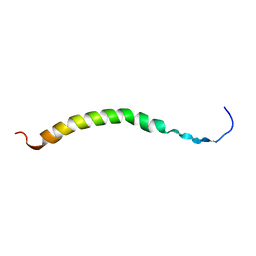 | | SOLUTION NMR STRUCTURES OF THE MAJOR COAT PROTEIN OF FILAMENTOUS BACTERIOPHAGE M13 SOLUBILIZED IN DODECYLPHOSPHOCHOLINE MICELLES, 25 LOWEST ENERGY STRUCTURES | | Descriptor: | M13 MAJOR COAT PROTEIN | | Authors: | Papavoine, C.H.M, Christiaans, B.E.C, Folmer, R.H.A, Konings, R.N.H, Hilbers, C.W. | | Deposit date: | 1998-04-16 | | Release date: | 1998-11-11 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the M13 major coat protein in detergent micelles: a basis for a model of phage assembly involving specific residues.
J.Mol.Biol., 282, 1998
|
|
1KG1
 
 | | NMR structure of the NIP1 elicitor protein from Rhynchosporium secalis | | Descriptor: | Necrosis Inducing Protein 1 | | Authors: | Van't Slot, K.A, Van den Burg, H.A, Kloks, C.P, Hilbers, C.W, Knogge, W, Papavoine, C.H. | | Deposit date: | 2001-11-26 | | Release date: | 2003-11-11 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Plant Disease Resistance-triggering Protein NIP1 from the Fungus Rhynchosporium secalis Shows a Novel beta-Sheet Fold.
J.Biol.Chem., 278, 2003
|
|
1A60
 
 | | NMR STRUCTURE OF A CLASSICAL PSEUDOKNOT: INTERPLAY OF SINGLE-AND DOUBLE-STRANDED RNA, 24 STRUCTURES | | Descriptor: | TYMV PSEUDOKNOT | | Authors: | Kolk, M.H, Van Der Graaf, M, Wijmenga, S.S, Pleij, C.W.A, Heus, H.A, Hilbers, C.W. | | Deposit date: | 1998-03-04 | | Release date: | 1998-05-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of a classical pseudoknot: interplay of single- and double-stranded RNA.
Science, 280, 1998
|
|
1YHB
 
 | | CRYSTAL STRUCTURES OF Y41H AND Y41F MUTANTS OF GENE V PROTEIN FROM FF PHAGE SUGGEST POSSIBLE PROTEIN-PROTEIN INTERACTIONS IN GVP-SSDNA COMPLEX | | Descriptor: | GENE V PROTEIN | | Authors: | Guan, Y, Zhang, H, Konings, R.N.H, Hilbers, C.W, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1994-04-14 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of Y41H and Y41F mutants of gene V protein from Ff phage suggest possible protein-protein interactions in the GVP-ssDNA complex.
Biochemistry, 33, 1994
|
|
1YHA
 
 | | CRYSTAL STRUCTURES OF Y41H AND Y41F MUTANTS OF GENE V PROTEIN FROM FF PHAGE SUGGEST POSSIBLE PROTEIN-PROTEIN INTERACTIONS IN GVP-SSDNA COMPLEX | | Descriptor: | GENE V PROTEIN | | Authors: | Guan, Y, Zhang, H, Konings, R.N.H, Hilbers, C.W, Terwilliger, T.C, Wang, A.H.-J. | | Deposit date: | 1994-04-14 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of Y41H and Y41F mutants of gene V protein from Ff phage suggest possible protein-protein interactions in the GVP-ssDNA complex.
Biochemistry, 33, 1994
|
|
1PFS
 
 | | SOLUTION NMR STRUCTURE OF THE SINGLE-STRANDED DNA BINDING PROTEIN OF THE FILAMENTOUS PSEUDOMONAS PHAGE PF3, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PF3 SINGLE-STRANDED DNA BINDING PROTEIN | | Authors: | Folmer, R.H.A, Nilges, M, Konings, R.N.H, Hilbers, C.W. | | Deposit date: | 1996-08-03 | | Release date: | 1997-02-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the single-stranded DNA binding protein of the filamentous Pseudomonas phage Pf3: similarity to other proteins binding to single-stranded nucleic acids.
EMBO J., 14, 1995
|
|
1N66
 
 | | Structure of the pyrimidine-rich internal loop in the Y-domain of poliovirus 3'UTR | | Descriptor: | internal loop in the Y-domain of poliovirus 3'UTR | | Authors: | Lescrinier, E.M, Tessari, M, van Kuppeveld, F.J, Melchers, W.J, Hilbers, C.W, Heus, H.A. | | Deposit date: | 2002-11-08 | | Release date: | 2003-08-19 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Pyrimidine-rich Internal Loop in the Poliovirus 3'-UTR: The Importance of Maintaining Pseudo-2-fold Symmetry in RNA Helices Containing Two Adjacent Non-canonical Base-pairs.
J.Mol.Biol., 331, 2003
|
|
1E95
 
 | | Solution structure of the pseudoknot of SRV-1 RNA, involved in ribosomal frameshifting | | Descriptor: | RNA (5'-(*GP*CP*GP*GP*CP*CP*AP*GP*CP*UP*CP* CP*AP*GP*GP*CP*CP*GP*CP*CP*AP*AP*AP*CP* AP*AP*UP*AP*UP*GP*GP*AP*GP*CP*AP*C)-3') | | Authors: | Michiels, P.J.A, Versleyen, A, Pleij, C.W.A, Hilbers, C.W, Heus, H.A. | | Deposit date: | 2000-10-09 | | Release date: | 2001-08-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Pseudoknot of Srv-1 RNA, Involved in Ribosomal Frameshifting
J.Mol.Biol., 310, 2001
|
|
1ATO
 
 | | THE STRUCTURE OF THE ISOLATED, CENTRAL HAIRPIN OF THE HDV ANTIGENOMIC RIBOZYME, NMR, 10 STRUCTURES | | Descriptor: | RNA (5'-R(*GP*GP*CP*AP*CP*CP*UP*CP*CP*UP*CP*GP*CP*GP*GP*UP*GP*CP*C)-3') | | Authors: | Kolk, M.H, Heus, H.A, Hilbers, C.W. | | Deposit date: | 1997-08-14 | | Release date: | 1997-11-12 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The structure of the isolated, central hairpin of the HDV antigenomic ribozyme: novel structural features and similarity of the loop in the ribozyme and free in solution.
EMBO J., 16, 1997
|
|
1B4Y
 
 | | STRUCTURE AND MECHANISM OF FORMATION OF THE H-Y5 ISOMER OF AN INTRAMOLECULAR DNA TRIPLE HELIX. | | Descriptor: | DNA (H-Y5 TRIPLE HELIX) | | Authors: | Van Dongen, M.J.P, Doreleijers, J.F, Van Der Marel, G.A, Van Boom, J.H, Hilbers, C.W, Wijmenga, S.S. | | Deposit date: | 1998-12-30 | | Release date: | 1999-09-13 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Structure and mechanism of formation of the H-y5 isomer of an intramolecular DNA triple helix.
Nat.Struct.Biol., 6, 1999
|
|
1AC7
 
 | | STRUCTURAL FEATURES OF THE DNA HAIRPIN D(ATCCTAGTTATAGGAT): THE FORMATION OF A G-A BASE PAIR IN THE LOOP, NMR, 10 STRUCTURES | | Descriptor: | DNA (5'-D(*AP*TP*CP*CP*TP*AP*GP*TP*TP*AP*TP*AP*GP*GP*AP*T)-3') | | Authors: | Van Dongen, M.J.P, Mooren, M.M.W, Willems, E.F.A, Van Der Marel, G.A, Van Boom, J.H, Wijmenga, S.S, Hilbers, C.W. | | Deposit date: | 1997-02-14 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural features of the DNA hairpin d(ATCCTA-GTTA-TAGGAT): formation of a G-A base pair in the loop.
Nucleic Acids Res., 25, 1997
|
|
1E4P
 
 | |
1H95
 
 | | Solution structure of the single-stranded DNA-binding Cold Shock Domain (CSD) of human Y-box protein 1 (YB1) determined by NMR (10 lowest energy structures) | | Descriptor: | Y-BOX BINDING PROTEIN | | Authors: | Kloks, C.P.A.M, Spronk, C.A.E.M, Hoffmann, A, Vuister, G.W, Grzesiek, S, Hilbers, C.W. | | Deposit date: | 2001-02-23 | | Release date: | 2002-02-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The Solution Structure and DNA-Binding Properties of the Cold-Shock Domain of the Human Y-Box Protein Yb-1.
J.Mol.Biol., 316, 2002
|
|
1EJZ
 
 | | SOLUTION STRUCTURE OF A HNA-RNA HYBRID | | Descriptor: | DNA (5'-H(*(6HG)P*(6HC)P*(6HG)P*(6HT)P*(6HA)P*(6HG)P*(6HC)P*(6HG))-3'), PHOSPHORIC ACID MONO-(3-HYDROXY-PROPYL) ESTER, RNA (5'-R(*CP*GP*CP*UP*AP*CP*GP*C)-3') | | Authors: | Lescrnier, E, Esnouf, R, Heus, H.A, Hilbers, C.W, Herdewijn, P. | | Deposit date: | 2000-03-06 | | Release date: | 2000-10-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a HNA-RNA hybrid.
Chem.Biol., 7, 2000
|
|
