5XEC
 
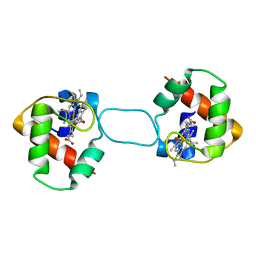 | | Heterodimer constructed from PA cyt c551-HT cyt c552 and HT cyt c552-PA cyt c551 chimeric proteins | | Descriptor: | Cytochrome c-551,Cytochrome c-552, Cytochrome c-552,Cytochrome c-551, HEME C | | Authors: | Zhang, M, Nakanishi, T, Yamanaka, M, Nagao, S, Yanagisawa, S, Shomura, Y, Shibata, N, Ogura, T, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-04-04 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Rational Design of Domain-Swapping-Based c-Type Cytochrome Heterodimers by Using Chimeric Proteins.
Chembiochem, 18, 2017
|
|
5XVC
 
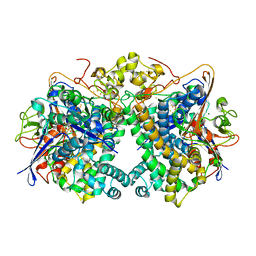 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in a ferricyanide-oxidized condition | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE3-S4 CLUSTER, FE4-S4-O CLUSTER, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
5XED
 
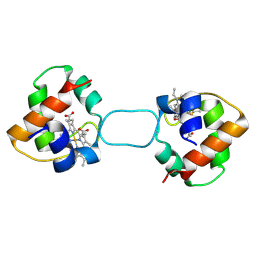 | | Heterodimer constructed from M61A PA cyt c551-HT cyt c552 and HT cyt c552-PA cyt c551 chimeric proteins | | Descriptor: | Cytochrome c-551,Cytochrome c-552, Cytochrome c-552,Cytochrome c-551, HEME C | | Authors: | Zhang, M, Nakanishi, T, Yamanaka, M, Nagao, S, Yanagisawa, S, Shomura, Y, Shibata, N, Ogura, T, Higuchi, Y, Hirota, S. | | Deposit date: | 2017-04-04 | | Release date: | 2017-08-09 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Rational Design of Domain-Swapping-Based c-Type Cytochrome Heterodimers by Using Chimeric Proteins.
Chembiochem, 18, 2017
|
|
5XVD
 
 | | [NiFe]-hydrogenase (Hyb-type) from Citrobacter sp. S-77 in an air-oxidized condition | | Descriptor: | FE3-S4 CLUSTER, FE4-S4-O CLUSTER, GLYCEROL, ... | | Authors: | Nishikawa, K, Matsuura, H, Muhd Noor, N.D, Tai, H, Hirota, S, Kim, J, Kang, J, Tateno, M, Yoon, K.S, Ogo, S, Shomura, Y, Higuchi, Y. | | Deposit date: | 2017-06-27 | | Release date: | 2018-06-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Redox-dependent conformational changes of a proximal [4Fe-4S] cluster in Hyb-type [NiFe]-hydrogenase to protect the active site from O2.
Chem.Commun.(Camb.), 54, 2018
|
|
5Y34
 
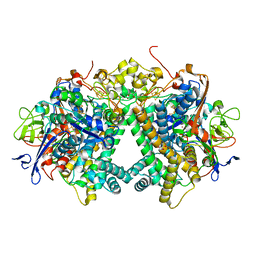 | | Membrane-bound respiratory [NiFe]-hydrogenase from Hydrogenovibrio marinus in a ferricyanide-oxidized condition | | Descriptor: | CARBONMONOXIDE-(DICYANO) IRON, FE3-S4 CLUSTER, FE4-S3 CLUSTER, ... | | Authors: | Shomura, Y, Yoon, K.S, Nishihara, H, Higuchi, Y. | | Deposit date: | 2017-07-27 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural basis for a [4Fe-3S] cluster in the oxygen-tolerant membrane-bound [NiFe]-hydrogenase
Nature, 479, 2011
|
|
3UX0
 
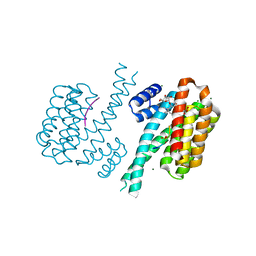 | | Crystal structure of human 14-3-3 sigma in complex with TASK-3 peptide and stabilizer Fusicoccin H | | Descriptor: | (4R,5R,6R,6aS,9S,9aE,10aR)-5-hydroxy-9-(hydroxymethyl)-6,10a-dimethyl-3-(propan-2-yl)-1,2,4,5,6,6a,7,8,9,10a-decahydrodicyclopenta[a,d][8]annulen-4-yl alpha-D-gulopyranoside, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Thiel, P, Bartel, M, Anders, C, Higuchi, Y, Schumacher, B, Kato, N, Ottmann, C. | | Deposit date: | 2011-12-03 | | Release date: | 2013-01-02 | | Last modified: | 2013-05-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of k(+) channels at the cell surface.
Chem.Biol., 20, 2013
|
|
3WC8
 
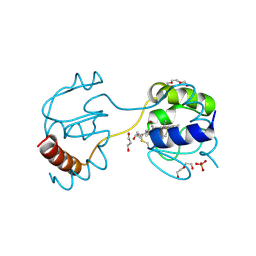 | | Dimeric horse cytochrome c obtained by refolding with desalting method | | Descriptor: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | Authors: | Parui, P.P, Deshpande, M.S, Nagao, S, Kamikubo, H, Komori, H, Higuchi, Y, Kataoka, M, Hirota, S. | | Deposit date: | 2013-05-25 | | Release date: | 2013-12-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Formation of Oligomeric Cytochrome c during Folding by Intermolecular Hydrophobic Interaction between N- and C-Terminal alpha-Helices
Biochemistry, 52, 2013
|
|
3WUI
 
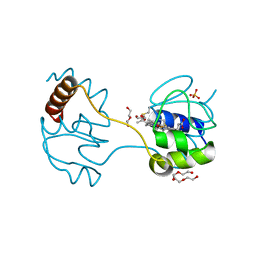 | | Dimeric horse cytochrome c formed by refolding from molten globule state | | Descriptor: | Cytochrome c, DI(HYDROXYETHYL)ETHER, HEME C, ... | | Authors: | Deshpande, M.S, Parui, P.P, Kamikubo, H, Yamanaka, M, Nagao, S, Komori, H, Kataoka, M, Higuchi, Y, Hirota, S. | | Deposit date: | 2014-04-25 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Formation of domain-swapped oligomer of cytochrome C from its molten globule state oligomer.
Biochemistry, 53, 2014
|
|
3A20
 
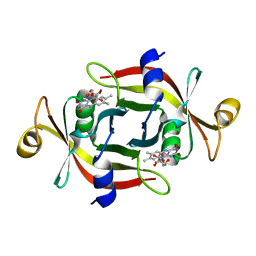 | |
3A8Q
 
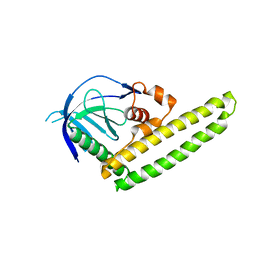 | | Low-resolution crystal structure of the Tiam2 PHCCEx domain | | Descriptor: | T-lymphoma invasion and metastasis-inducing protein 2 | | Authors: | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
3A8P
 
 | | Crystal structure of the Tiam2 PHCCEx domain | | Descriptor: | T-lymphoma invasion and metastasis-inducing protein 2 | | Authors: | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
3A8N
 
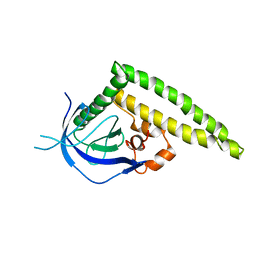 | | Crystal structure of the Tiam1 PHCCEx domain | | Descriptor: | T-lymphoma invasion and metastasis-inducing protein 1 | | Authors: | Terawaki, S, Kitano, K, Mori, T, Zhai, Y, Higuchi, Y, Itoh, N, Watanabe, T, Kaibuchi, K, Hakoshima, T. | | Deposit date: | 2009-10-07 | | Release date: | 2009-11-24 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (4.5 Å) | | Cite: | The PHCCEx domain of Tiam1/2 is a novel protein- and membrane-binding module
Embo J., 29, 2010
|
|
5XYO
 
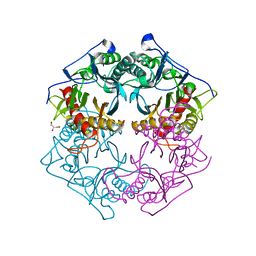 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122G mutant | | Descriptor: | CHLORIDE ION, Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, ... | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5XYP
 
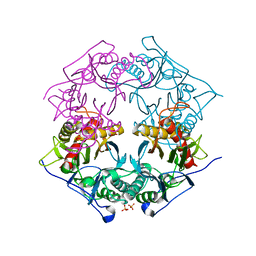 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122R mutant | | Descriptor: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, PHOSPHATE ION | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5XYG
 
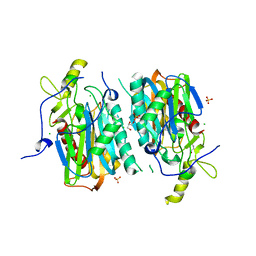 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72. | | Descriptor: | CHLORIDE ION, Endotype 6-aminohexanoat-oligomer hydrolase, GLYCEROL, ... | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-07 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5XYS
 
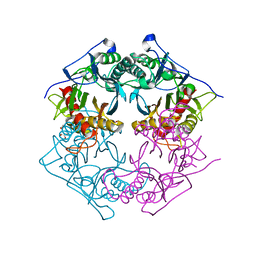 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122V mutant | | Descriptor: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, PHOSPHATE ION | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5Y0L
 
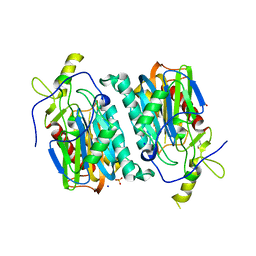 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122G/H130Y mutant | | Descriptor: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, SODIUM ION, ... | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-18 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.385 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
5Y0M
 
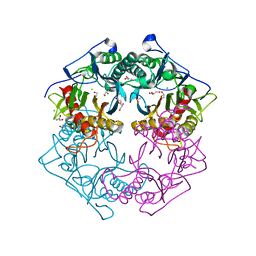 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D36A/D122G/H130Y/E263Q mutant | | Descriptor: | CHLORIDE ION, Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, ... | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-18 | | Release date: | 2018-07-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
2Z1T
 
 | |
5XYQ
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., D122K mutant | | Descriptor: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, PHOSPHATE ION | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
2Z1U
 
 | |
5XYT
 
 | | Structure of 6-aminohexanoate-oligomer hydrolase from Arthrobacter sp. KI72., H130Y mutant | | Descriptor: | Endo-type 6-aminohexanoate oligomer hydrolase, GLYCEROL, SULFATE ION | | Authors: | Negoro, S, Shibata, N, Nagai, K, Higuchi, Y. | | Deposit date: | 2017-07-10 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of the correct subunit assembly, aggregation, and intracellular degradation of nylon hydrolase
Sci Rep, 8, 2018
|
|
3X39
 
 | | Domain-swapped dimer of Pseudomonas aeruginosa cytochrome c551 | | Descriptor: | Cytochrome c-551, HEME C | | Authors: | Nagao, S, Ueda, M, Osuka, H, Komori, H, Kamikubo, H, Kataoka, M, Higuchi, Y, Hirota, S. | | Deposit date: | 2015-01-16 | | Release date: | 2015-04-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Domain-Swapped Dimer of Pseudomonas aeruginosa Cytochrome c551: Structural Insights into Domain Swapping of Cytochrome c Family Proteins
Plos One, 10, 2015
|
|
2ZM8
 
 | | Structure of 6-Aminohexanoate-dimer Hydrolase, S112A/D370Y Mutant Complexed with 6-Aminohexanoate-dimer | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-AMINOHEXANOIC ACID, 6-aminohexanoate-dimer hydrolase, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Negoro, S. | | Deposit date: | 2008-04-14 | | Release date: | 2009-04-14 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
2ZM2
 
 | | Structure of 6-aminohexanoate-dimer hydrolase, A61V/A124V/R187S/F264C/G291R/G338A/D370Y mutant (Hyb-S4M94) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 6-aminohexanoate-dimer hydrolase, GLYCEROL, ... | | Authors: | Ohki, T, Shibata, N, Higuchi, Y, Kawashima, Y, Takeo, M, Kato, D, Nego, S. | | Deposit date: | 2008-04-10 | | Release date: | 2009-04-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Two alternative modes for optimizing nylon-6 byproduct hydrolytic activity from a carboxylesterase with a beta-lactamase fold: X-ray crystallographic analysis of directly evolved 6-aminohexanoate-dimer hydrolase.
Protein Sci., 18, 2009
|
|
