2XON
 
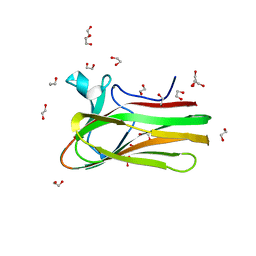 | | Structure of TmCBM61 in complex with beta-1,4-galactotriose at 1.4 A resolution | | Descriptor: | 1,2-ETHANEDIOL, ARABINOGALACTAN ENDO-1,4-BETA-GALACTOSIDASE, CALCIUM ION, ... | | Authors: | Cid, M, Lodberg-Pedersen, H, Kaneko, S, Coutinho, P.M, Henrissat, B, Willats, W.G.T, Boraston, A.B. | | Deposit date: | 2010-08-20 | | Release date: | 2010-09-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Recognition of the Helical Structure of Beta-1,4-Galactan by a New Family of Carbohydrate-Binding Modules.
J.Biol.Chem., 285, 2010
|
|
5MUM
 
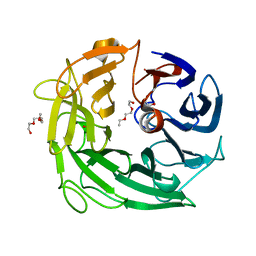 | | Glycoside Hydrolase BACINT_00347 | | Descriptor: | 1,2-ETHANEDIOL, BACINT_00347, PENTAETHYLENE GLYCOL | | Authors: | Munoz-Munoz, J, Cartmell, A, Terrapon, N, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2017-01-13 | | Release date: | 2017-04-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unusual active site location and catalytic apparatus in a glycoside hydrolase family.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5A8Q
 
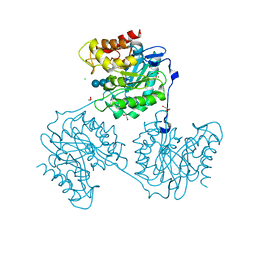 | | Crystal structure beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A obtained by soaking | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sulzenbacher, G, Lafond, M, Freyd, T, Henrissat, B, Coutinho, R.M, Berrin, J.G, Garron, M.L. | | Deposit date: | 2015-07-16 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Quaternary Structure of a Glycoside Hydrolase Dictates Specificity Towards Beta-Glucans
J.Biol.Chem., 291, 2016
|
|
1UXZ
 
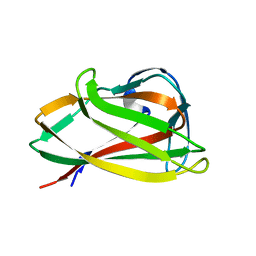 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A | | Descriptor: | CELLULASE B | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
5A8P
 
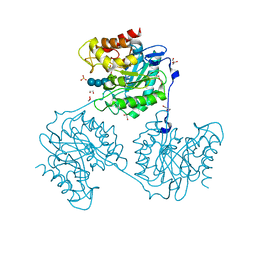 | | Crystal structure beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide B | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sulzenbacher, G, Lafond, M, Freyd, T, Henrissat, B, Coutinho, R.M, Berrin, J.G, Garron, M.L. | | Deposit date: | 2015-07-16 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Quaternary Structure of a Glycoside Hydrolase Dictates Specificity Towards Beta-Glucans
J.Biol.Chem., 291, 2016
|
|
5A94
 
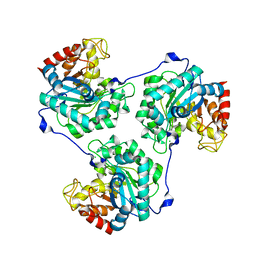 | | Crystal structure of beta-Glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 1 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, PUTATIVE RETAINING B-GLYCOSIDASE, ... | | Authors: | Sulzenbacher, G, Lafond, M, Freyd, T, Henrissat, B, Coutinho, R.M, Berrin, J.G, Garron, M.L. | | Deposit date: | 2015-07-17 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | The Quaternary Structure of a Glycoside Hydrolase Dictates Specificity Towards Beta-Glucans
J.Biol.Chem., 291, 2016
|
|
5A95
 
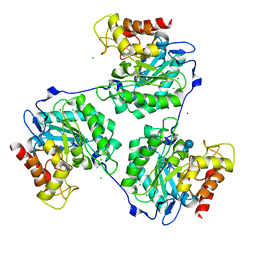 | | Crystal structure of beta-glucanase SdGluc5_26A from Saccharophagus degradans in complex with tetrasaccharide A, form 2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Sulzenbacher, G, Lafond, M, Freyd, T, Henrissat, B, Coutinho, R.M, Berrin, J.G, Garron, M.L. | | Deposit date: | 2015-07-17 | | Release date: | 2016-01-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The Quaternary Structure of a Glycoside Hydrolase Dictates Specificity Towards Beta-Glucans
J.Biol.Chem., 291, 2016
|
|
1UYX
 
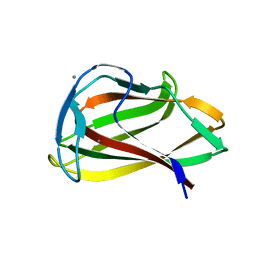 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with cellobiose | | Descriptor: | CALCIUM ION, CELLULASE B, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
1UZ0
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with Glc-4Glc-3Glc-4Glc | | Descriptor: | CALCIUM ION, CELLULASE B, CHLORIDE ION, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
5MUL
 
 | | Glycoside Hydrolase BT3686 bound to Glucuronic Acid | | Descriptor: | Neuraminidase, beta-D-glucopyranuronic acid | | Authors: | Munoz-Munoz, J, Cartmell, A, Terrapon, N, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2017-01-13 | | Release date: | 2017-04-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Unusual active site location and catalytic apparatus in a glycoside hydrolase family.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
1E1E
 
 | | Crystal structure of a Monocot (Maize ZMGlu1) beta-glucosidase | | Descriptor: | BETA-GLUCOSIDASE | | Authors: | Czjzek, M, Cicek, M, Bevan, D.R, Henrissat, B, Esen, A. | | Deposit date: | 2000-05-03 | | Release date: | 2001-02-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of a Monocotyledon (Maize Zmglu1) Beta-Glucosidase and a Model of its Complex with P-Nitrophenyl Beta-D-Thioglucoside
Biochem.J., 354, 2001
|
|
5NOK
 
 | | Polysaccharide Lyase BACCELL_00875 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BACCELL_00875 | | Authors: | Cartmell, A, Munoz-Munoz, J, Terrapon, N, Basle, A, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2017-04-12 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | An evolutionarily distinct family of polysaccharide lyases removes rhamnose capping of complex arabinogalactan proteins.
J. Biol. Chem., 292, 2017
|
|
5A8M
 
 | | Crystal structure of the selenomethionine derivative of beta-glucanase SdGluc5_26A from Saccharophagus degradans | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sulzenbacher, G, Lafond, M, Freyd, T, Henrissat, B, Coutinho, R.M, Berrin, J.G, Garron, M.L. | | Deposit date: | 2015-07-16 | | Release date: | 2016-01-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | The Quaternary Structure of a Glycoside Hydrolase Dictates Specificity Towards Beta-Glucans
J.Biol.Chem., 291, 2016
|
|
1G1K
 
 | | COHESIN MODULE FROM THE CELLULOSOME OF CLOSTRIDIUM CELLULOLYTICUM | | Descriptor: | SCAFFOLDING PROTEIN | | Authors: | Spinelli, S, Fierobe, H.-P, Belaich, A, Belaich, J.-P, Henrissat, B, Cambillau, C. | | Deposit date: | 2000-10-12 | | Release date: | 2000-11-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a cohesin module from Clostridium cellulolyticum: implications for dockerin recognition.
J.Mol.Biol., 304, 2000
|
|
1UYP
 
 | | The three-dimensional structure of beta-fructosidase (invertase) from Thermotoga maritima | | Descriptor: | BETA-FRUCTOSIDASE, CITRIC ACID, GLYCEROL, ... | | Authors: | Alberto, F, Bignon, C, Sulzenbacher, G, Henrissat, B, Czjzek, M. | | Deposit date: | 2004-03-02 | | Release date: | 2004-03-22 | | Last modified: | 2018-06-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The three-dimensional structure of invertase (beta-fructosidase) from Thermotoga maritima reveals a bimodular arrangement and an evolutionary relationship between retaining and inverting glycosidases.
J. Biol. Chem., 279, 2004
|
|
1UY0
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with glc-1,3-glc-1,4-glc-1,3-glc | | Descriptor: | CALCIUM ION, CELLULASE B, CHLORIDE ION, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-01 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
1URX
 
 | | Crystallographic structure of beta-agarase A in complex with oligoagarose | | Descriptor: | 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-alpha-D-galactopyranose, 3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose-(1-3)-beta-D-galactopyranose-(1-4)-3,6-anhydro-alpha-L-galactopyranose, BETA-AGARASE A, ... | | Authors: | Allouch, J, Helbert, W, Henrissat, B, Czjzek, M. | | Deposit date: | 2003-11-12 | | Release date: | 2004-03-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Parallel Substrate Binding Sites in a Beta-Agarase Suggest a Novel Mode of Action on Double-Helical Agarose
Structure, 12, 2004
|
|
1UYZ
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with xylotetraose | | Descriptor: | CALCIUM ION, CELLULASE B, GLYCEROL, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
1UYY
 
 | | Carbohydrate binding module (CBM6cm-2) from Cellvibrio mixtus lichenase 5A in complex with cellotriose | | Descriptor: | CALCIUM ION, CELLULASE B, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose, ... | | Authors: | Czjzek, M, Pires, V.M.R, Henshaw, J, Prates, J.A.M, Bolam, D, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2004-03-03 | | Release date: | 2004-03-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | The Crystal Structure of the Family 6 Carbohydrate Binding Module from Cellvibrio Mixtus Endoglucanase 5A in Complex with Oligosaccharides Reveals Two Distinct Binding Sites with Different Ligand Specificities
J.Biol.Chem., 279, 2004
|
|
1V02
 
 | | Crystal structure of the Sorghum bicolor dhurrinase 1 | | Descriptor: | DHURRINASE | | Authors: | Moriniere, J, Verdoucq, L, Bevan, D.R, Esen, A, Henrissat, B, Czjzek, M. | | Deposit date: | 2004-03-22 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Determinants of Substrate Specificity in Family 1 Beta-Glucosidases: Novel Insights from the Crystal Structure of Sorghum Dhurrinase-1, a Plant Beta-Glucosidase with Strict Specificity, in Complex with its Natural Substrate
J.Biol.Chem., 279, 2004
|
|
1V03
 
 | | Crystal structure of the Sorghum bicolor dhurrinase 1 | | Descriptor: | ACETONITRILE, DHURRINASE, PHENOL, ... | | Authors: | Moriniere, J, Verdoucq, L, Bevan, D.R, Esen, A, Henrissat, B, Czjzek, M. | | Deposit date: | 2004-03-22 | | Release date: | 2004-05-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Determinants of Substrate Specificity in Family 1 Beta-Glucosidases: Novel Insights from the Crystal Structure of Sorghum Dhurrinase-1, a Plant Beta-Glucosidase with Strict Specificity, in Complex with its Natural Substrate
J.Biol.Chem., 279, 2004
|
|
1V08
 
 | | Crystal structure of the Zea maze beta-glucosidase-1 in complex with gluco-tetrazole | | Descriptor: | BETA-GLUCOSIDASE, NOJIRIMYCINE TETRAZOLE | | Authors: | Moriniere, J, Verdoucq, L, Bevan, D.R, Esen, A, Henrissat, B, Czjzek, M. | | Deposit date: | 2004-03-25 | | Release date: | 2004-05-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Determinants of Substrate Specificity in Family 1 Beta-Glucosidases: Novel Insights from the Crystal Structure of Sorghum Dhurrinase-1, a Plant Beta-Glucosidase with Strict Specificity, in Complex with its Natural Substrate
J.Biol.Chem., 279, 2004
|
|
1E4N
 
 | | Crystal structure of the inactive mutant Monocot (Maize ZMGlu1) beta-glucosidase ZMGluE191D in complex with the natural aglycone DIMBOA | | Descriptor: | 2,4-DIHYDROXY-7-(METHYLOXY)-2H-1,4-BENZOXAZIN-3(4H)-ONE, BETA-GLUCOSIDASE | | Authors: | Czjzek, M, Cicek, M, Bevan, D.R, Zamboni, V, Henrissat, B, Esen, A. | | Deposit date: | 2000-07-11 | | Release date: | 2000-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Mechanism of Substrate (Aglycone) Specificity in Beta -Glucosidases is Revealed by Crystal Structures of Mutant Maize Beta -Glucosidase-Dimboa, -Dimboaglc, and -Dhurrin Complexes
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
1E4L
 
 | | Crystal structure of the inactive mutant Monocot (Maize ZMGlu1) beta-glucosidase ZM Glu191Asp | | Descriptor: | BETA-GLUCOSIDASE, CHLOROPLASTIC, GLYCEROL | | Authors: | Czjzek, M, Cicek, M, Bevan, D.R, Henrissat, B, Esen, A. | | Deposit date: | 2000-07-10 | | Release date: | 2000-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Mechanism of Substrate (Aglycone) Specificity in Beta -Glucosidases is Revealed by Crystal Structures of Mutant Maize Beta -Glucosidase-Dimboa, -Dimboaglc, and -Dhurrin Complexes
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
5NO8
 
 | | Polysaccharide Lyase BACCELL_00875 | | Descriptor: | BACCELL_00875, GLYCEROL | | Authors: | Cartmell, A, Munoz-Munoz, J, Terrapon, N, Basle, A, Henrissat, B, Gilbert, H.J. | | Deposit date: | 2017-04-11 | | Release date: | 2017-06-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | An evolutionarily distinct family of polysaccharide lyases removes rhamnose capping of complex arabinogalactan proteins.
J. Biol. Chem., 292, 2017
|
|
