3MJE
 
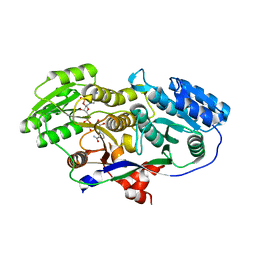 | | Structure of A-type Ketoreductases from Modular Polyketide Synthase | | Descriptor: | AmphB, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, J, Taylor, C.A, Piasecki, S.K, Keatinge-Clay, A.T. | | Deposit date: | 2010-04-12 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Structural and Functional Analysis of A-Type Ketoreductases from the Amphotericin Modular Polyketide Synthase.
Structure, 18, 2010
|
|
3MJT
 
 | | Structure of A-type Ketoreductases from Modular Polyketide Synthase | | Descriptor: | AmphB, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, J, Taylor, C.A, Piasecki, S.K, Keatinge-Clay, A.T. | | Deposit date: | 2010-04-13 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Analysis of A-Type Ketoreductases from the Amphotericin Modular Polyketide Synthase.
Structure, 18, 2010
|
|
3MJV
 
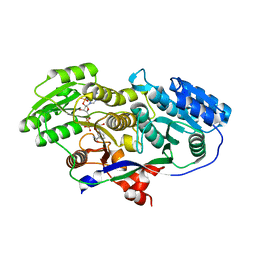 | | Structure of A-type Ketoreductases from Modular Polyketide Synthase | | Descriptor: | AmphB, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, J, Taylor, C.A, Piasecki, S.K, Keatinge-Clay, A.T. | | Deposit date: | 2010-04-13 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural and Functional Analysis of A-Type Ketoreductases from the Amphotericin Modular Polyketide Synthase.
Structure, 18, 2010
|
|
4DIF
 
 | | Structure of A1-type ketoreductase | | Descriptor: | AmphB, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, J, Keatinge-Clay, A.T. | | Deposit date: | 2012-01-30 | | Release date: | 2013-02-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.521 Å) | | Cite: | Structure and mutagenesis of A2-type ketoreductase from modular polyketide synthase reveals insights into stereospecificity
To be Published
|
|
6NKO
 
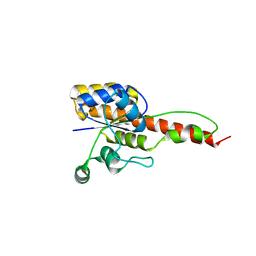 | | Crystal structure of ForH | | Descriptor: | ForH | | Authors: | Zheng, J, Irani, S, Zhang, Y. | | Deposit date: | 2019-01-07 | | Release date: | 2019-04-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.403 Å) | | Cite: | Identification of the Formycin A Biosynthetic Gene Cluster from Streptomyces kaniharaensis Illustrates the Interplay between Biological Pyrazolopyrimidine Formation and de Novo Purine Biosynthesis.
J. Am. Chem. Soc., 141, 2019
|
|
7VWA
 
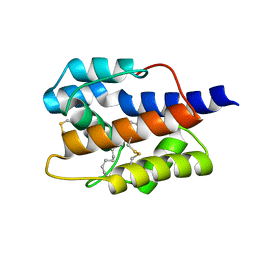 | |
7VW8
 
 | |
7VW9
 
 | |
3LTU
 
 | | 5-SeMe-dU containing DNA 8mer | | Descriptor: | 5'-D(*GP*(UMS)P*GP*(T5S)P*AP*CP*AP*C)-3' | | Authors: | Sheng, J, Hassan, A.E.A, Zhang, W, Gan, J, Huang, Z. | | Deposit date: | 2010-02-16 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Synthesis of Pyrimidine Modified Seleno-DNA as a Novel Approach to Antisense Candidate
Chemistryselect, 8, 2023
|
|
3IKI
 
 | | 5-SMe-dU containing DNA octamer | | Descriptor: | 5'-D(*GP*(UMS)P*GP*(US2)P*AP*CP*AP*C)-3', MAGNESIUM ION | | Authors: | Sheng, J, Hassan, A.E.A, Zhang, W, Gan, J, Huang, Z. | | Deposit date: | 2009-08-05 | | Release date: | 2010-03-09 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Hydrogen bond formation between the naturally modified nucleobase and phosphate backbone.
Nucleic Acids Res., 40, 2012
|
|
3IJN
 
 | |
3QP9
 
 | |
5E36
 
 | |
3FA1
 
 | |
4MSB
 
 | | RNA 10mer duplex with two 2'-5'-linkages | | Descriptor: | RNA 10mer duplex with two 2'-5'-linkages, STRONTIUM ION | | Authors: | Sheng, J, Li, L, Engelhart, A.E, Gan, J, Wang, J, Szostak, J.W. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into the effects of 2'-5' linkages on the RNA duplex.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7XNY
 
 | |
7XNX
 
 | |
7CTE
 
 | | Human Origin Recognition Complex, ORC2-5 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Origin recognition complex subunit 2, Origin recognition complex subunit 3, ... | | Authors: | Cheng, J, Li, N, Wang, X, Hu, J, Zhai, Y, Gao, N. | | Deposit date: | 2020-08-18 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insight into the assembly and conformational activation of human origin recognition complex.
Cell Discov, 6, 2020
|
|
7CTF
 
 | | Human origin recognition complex 1-5 State II | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Origin recognition complex subunit 1, Origin recognition complex subunit 2, ... | | Authors: | Cheng, J, Li, N, Wang, X, Hu, J, Zhai, Y, Gao, N. | | Deposit date: | 2020-08-18 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural insight into the assembly and conformational activation of human origin recognition complex.
Cell Discov, 6, 2020
|
|
5C56
 
 | | Crystal structure of USP7/HAUSP in complex with ICP0 | | Descriptor: | Ubiquitin E3 ligase ICP0, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Cheng, J, Li, Z, Gong, R, Fang, J, Yang, Y, Sun, C, Yang, H, Xu, Y. | | Deposit date: | 2015-06-19 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.685 Å) | | Cite: | Molecular mechanism for the substrate recognition of USP7.
Protein Cell, 6, 2015
|
|
4MS9
 
 | | Native RNA-10mer Structure: ccggcgccgg | | Descriptor: | Native RNA duplex 10mer, STRONTIUM ION | | Authors: | Sheng, J, Li, L, Engelhart, A.E, Gan, J, Wang, J, Szostak, J.W. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Structural insights into the effects of 2'-5' linkages on the RNA duplex.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
7CTG
 
 | | Human Origin Recognition Complex, ORC1-5 State I | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Origin recognition complex subunit 1, Origin recognition complex subunit 2, ... | | Authors: | Cheng, J, Li, N, Wang, X, Hu, J, Zhai, Y, Gao, N. | | Deposit date: | 2020-08-18 | | Release date: | 2021-01-06 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural insight into the assembly and conformational activation of human origin recognition complex.
Cell Discov, 6, 2020
|
|
4MSR
 
 | | RNA 10mer duplex with six 2'-5'-linkages | | Descriptor: | RNA 10mer duplex with six 2'-5'-linkages, STRONTIUM ION | | Authors: | Sheng, J, Li, L, Engelhart, A.E, Gan, J, Wang, J, Szostak, J.W. | | Deposit date: | 2013-09-18 | | Release date: | 2014-02-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural insights into the effects of 2'-5' linkages on the RNA duplex.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5B5H
 
 | | Hydrophobic ice-binding site confer hyperactivity on antifreeze protein from a snow mold fungus | | Descriptor: | Antifreeze protein, SODIUM ION, SULFATE ION | | Authors: | Cheng, J, Hanada, Y, Miura, A, Tsuda, S, Kondo, H. | | Deposit date: | 2016-05-06 | | Release date: | 2016-09-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Hydrophobic ice-binding sites confer hyperactivity of an antifreeze protein from a snow mold fungus.
Biochem.J., 473, 2016
|
|
7V3U
 
 | | Cryo-EM structure of MCM double hexamer with structured Mcm4-NSD | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, DNA replication licensing factor MCM2, DNA replication licensing factor MCM3, ... | | Authors: | Cheng, J, Li, N, Huo, Y, Dang, S, Tye, B, Gao, N, Zhai, Y. | | Deposit date: | 2021-08-11 | | Release date: | 2022-04-13 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural Insight into the MCM double hexamer activation by Dbf4-Cdc7 kinase.
Nat Commun, 13, 2022
|
|
