4FDU
 
 | | Crystal Structure of a Multiple Inositol Polyphosphate Phosphatase | | Descriptor: | 1,2-ETHANEDIOL, D-MYO-INOSITOL-HEXASULPHATE, Putative multiple inositol polyphosphate histidine phosphatase 1 | | Authors: | Li, A.W.H, Hemmings, A.M. | | Deposit date: | 2012-05-29 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.293 Å) | | Cite: | A Bacterial Homolog of a Eukaryotic Inositol Phosphate Signaling Enzyme Mediates Cross-kingdom Dialog in the Mammalian Gut.
Cell Rep, 6, 2014
|
|
4FDT
 
 | | Crystal Structure of a Multiple Inositol Polyphosphate Phosphatase | | Descriptor: | 1,2-ETHANEDIOL, PHOSPHATE ION, Putative multiple inositol polyphosphate histidine phosphatase 1 | | Authors: | Li, A.W.H, Hemmings, A.M. | | Deposit date: | 2012-05-29 | | Release date: | 2014-01-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A Bacterial Homolog of a Eukaryotic Inositol Phosphate Signaling Enzyme Mediates Cross-kingdom Dialog in the Mammalian Gut.
Cell Rep, 6, 2014
|
|
9EQH
 
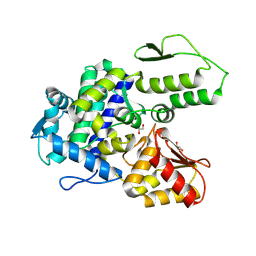 | | WWP2 WW2-2,3-linker-HECT (WWP2-LH) | | Descriptor: | GLYCEROL, Isoform 2 of NEDD4-like E3 ubiquitin-protein ligase WWP2, SODIUM ION | | Authors: | Dudey, A.P, Hemmings, A.M. | | Deposit date: | 2024-03-21 | | Release date: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Expanding the Inhibitor Space of the WWP1 and WWP2 HECT E3 Ligases
To Be Published
|
|
8OXE
 
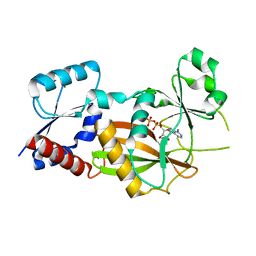 | | Inositol 1,3,4-trisphosphate 5/6-kinase 1 from Solanum tuberosum (StITPK1) in complex with ADP/Mg2+ | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Inositol-tetrakisphosphate 1-kinase, MAGNESIUM ION | | Authors: | Faba-Rodriguez, R, Li, A.W.H, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2023-05-02 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Crystal Structure and Enzymology of Solanum tuberosum Inositol Tris/Tetrakisphosphate Kinase 1 ( St ITPK1).
Biochemistry, 63, 2024
|
|
7R5Y
 
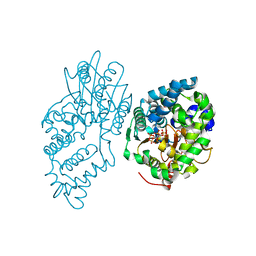 | |
7PUP
 
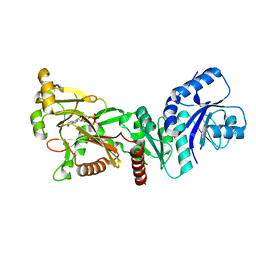 | | INOSITOL 1,3,4-TRISPHOSPHATE 5/6-KINASE 4 from Arabidopsis thaliana (AtITPK4) in complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Inositol 1,3,4-trisphosphate 5/6-kinase 4 | | Authors: | Whitfield, H, He, S, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2021-09-30 | | Release date: | 2022-10-12 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Diversification in the inositol tris/tetrakisphosphate kinase (ITPK) family: crystal structure and enzymology of the outlier AtITPK4.
Biochem.J., 480, 2023
|
|
5FD6
 
 | |
4XKT
 
 | | E coli BFR variant Y149F | | Descriptor: | Bacterioferritin, SULFATE ION | | Authors: | Bradley, J.M, Hemmings, A.M, Le Brun, N.E. | | Deposit date: | 2015-01-12 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Three Aromatic Residues are Required for Electron Transfer during Iron Mineralization in Bacterioferritin.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6GFH
 
 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with neo-IP5 and ATP | | Descriptor: | 1,2-ETHANEDIOL, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Whitfield, H.L, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-04-30 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A Fluorescent Probe Identifies Active Site Ligands of Inositol Pentakisphosphate 2-Kinase.
J. Med. Chem., 61, 2018
|
|
3TOR
 
 | | Crystal structure of Escherichia coli NrfA with Europium bound | | Descriptor: | CALCIUM ION, Cytochrome c nitrite reductase, EUROPIUM ION, ... | | Authors: | Lockwood, C.W.J, Clarke, T.A, Butt, J.N, Hemmings, A.M, Richardson, D.J. | | Deposit date: | 2011-09-06 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characterization of the active site and calcium binding in cytochrome c nitrite reductases.
Biochem.Soc.Trans., 39, 2011
|
|
3I9Z
 
 | | Crystal structure of a metallochaperone with a trinuclear Cu(I) cluster | | Descriptor: | COPPER (I) ION, Copper chaperone copZ | | Authors: | Hearnshaw, S.J, Zhou, L, Le Brun, N.E, Hemmings, A.M. | | Deposit date: | 2009-07-13 | | Release date: | 2009-11-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic insights into Cu(I) cluster transfer between the chaperone CopZ and its cognate Cu(I)-transporting P-type ATPase, CopA.
Biochem.J., 424, 2009
|
|
3L1T
 
 | | E. coli NrfA sulfite ocmplex | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Clarke, T.A, Hemmings, A.M, Butt, J.N. | | Deposit date: | 2009-12-14 | | Release date: | 2010-10-27 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Kinetic and thermodynamic resolution of the interactions between sulfite and the pentahaem cytochrome NrfA from Escherichia coli
Biochem.J., 431, 2010
|
|
5CU7
 
 | |
5CX4
 
 | |
3PHV
 
 | | X-RAY ANALYSIS OF HIV-1 PROTEINASE AT 2.7 ANGSTROMS RESOLUTION CONFIRMS STRUCTURAL HOMOLOGY AMONG RETROVIRAL ENZYMES | | Descriptor: | UNLIGANDED HIV-1 PROTEASE | | Authors: | Lapatto, R, Blundell, T.L, Hemmings, A, Wilderspin, A, Wood, S.P, Danley, D.E, Geoghegan, K.F, Hawrylik, S.J, Hobart, P.M. | | Deposit date: | 1991-11-04 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray analysis of HIV-1 proteinase at 2.7 A resolution confirms structural homology among retroviral enzymes.
Nature, 342, 1989
|
|
7Z3V
 
 | | Escherichia coli periplasmic phytase AppA D304E mutant, complex with myo-inositol hexakissulfate | | Descriptor: | Acidphosphatase, D-MYO-INOSITOL-HEXASULPHATE, POTASSIUM ION | | Authors: | Acquistapace, I.M, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2022-03-02 | | Release date: | 2022-06-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA.
Int J Mol Sci, 23, 2022
|
|
7Z2Y
 
 | | Escherichia coli periplasmic phytase AppA T305E mutant, complex with myo-inositol hexakissulfate | | Descriptor: | Acidphosphatase, D-MYO-INOSITOL-HEXASULPHATE, NICKEL (II) ION | | Authors: | Acquistapace, I.M, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2022-03-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA.
Int J Mol Sci, 23, 2022
|
|
7Z2W
 
 | | Escherichia coli periplasmic phytase AppA D304A,T305E mutant, complex with myo-inositol hexakissulfate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acidphosphatase, D-MYO-INOSITOL-HEXASULPHATE, ... | | Authors: | Acquistapace, I.M, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2022-03-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA.
Int J Mol Sci, 23, 2022
|
|
7Z1J
 
 | | Escherichia coli periplasmic phytase AppA, complex with phosphate | | Descriptor: | Acidphosphatase, MAGNESIUM ION, NICKEL (II) ION, ... | | Authors: | Acquistapace, I.M, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2022-02-24 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA.
Int J Mol Sci, 23, 2022
|
|
7Z2S
 
 | | Escherichia coli periplasmic phytase AppA, complex with myo-inositol hexakissulfate | | Descriptor: | Acidphosphatase, D-MYO-INOSITOL-HEXASULPHATE, NICKEL (II) ION, ... | | Authors: | Acquistapace, I.M, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2022-02-28 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA.
Int J Mol Sci, 23, 2022
|
|
7Z32
 
 | | Escherichia coli periplasmic phytase AppA D304A mutant, phosphohistidine intermediate | | Descriptor: | Acidphosphatase, NICKEL (II) ION | | Authors: | Acquistapace, I.M, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2022-03-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA.
Int J Mol Sci, 23, 2022
|
|
7Z2T
 
 | | Escherichia coli periplasmic phytase AppA D304A mutant, complex with myo-inositol hexakissulfate | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Acidphosphatase, D-MYO-INOSITOL-HEXASULPHATE, ... | | Authors: | Acquistapace, I.M, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2022-02-28 | | Release date: | 2022-03-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Insights to the Structural Basis for the Stereospecificity of the Escherichia coli Phytase, AppA.
Int J Mol Sci, 23, 2022
|
|
7ZGF
 
 | |
7ZGH
 
 | |
7ZGG
 
 | |
