1QO8
 
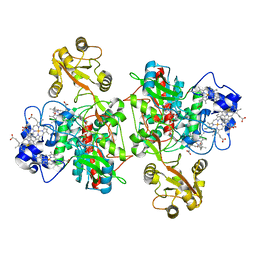 | | The structure of the open conformation of a flavocytochrome c3 fumarate reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, FLAVOCYTOCHROME C3 FUMARATE REDUCTASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Bamford, V, Dobbin, P.S, Richardson, D.J, Hemmings, A.M. | | Deposit date: | 1999-11-04 | | Release date: | 2000-11-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Open Conformation of a Flavocytochrome C3 Fumarate Reductase.
Nat.Struct.Biol., 6, 1999
|
|
4MT5
 
 | | Crystal structure of Mub-RV | | Descriptor: | Mucus binding proteinn | | Authors: | Etzold, S, Juge, N, Hemmings, A.M. | | Deposit date: | 2013-09-19 | | Release date: | 2014-08-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for adaptation of lactobacilli to gastrointestinal mucus.
ENVIRON.MICROBIOL., 16, 2014
|
|
4NG0
 
 | |
2RDZ
 
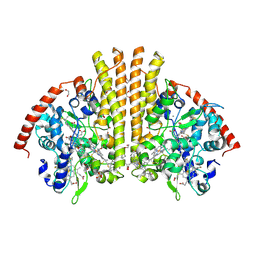 | | High Resolution Crystal Structure of the Escherichia coli Cytochrome c Nitrite Reductase. | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Clarke, T.A, Hemmings, A.M, RIchardson, D.J. | | Deposit date: | 2007-09-25 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Role of a Conserved Glutamine Residue in Tuning the Catalytic Activity of Escherichia coli Cytochrome c Nitrite Reductase.
Biochemistry, 47, 2008
|
|
2RF7
 
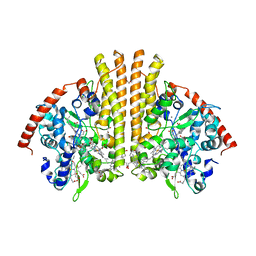 | | Crystal structure of the escherichia coli nrfa mutant Q263E | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Cytochrome c-552, ... | | Authors: | Clarke, T.A, Richardson, D.J, Hemmings, A.M. | | Deposit date: | 2007-09-28 | | Release date: | 2008-03-25 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Role of a Conserved Glutamine Residue in Tuning the Catalytic Activity of Escherichia coli Cytochrome c Nitrite Reductase.
Biochemistry, 47, 2008
|
|
6RXD
 
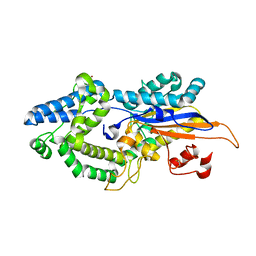 | |
6RXF
 
 | |
6RXG
 
 | |
6RXE
 
 | |
6SRR
 
 | |
6SQU
 
 | | Crystal structure of human SHIP2 catalytic domain in complex with 1,2,4 Dimer | | Descriptor: | 5,5'-(ethane-1,2-diylbis(oxy))bis(benzene-5,4,2,1,-tetrayl)hexakisphosphate, Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 2 | | Authors: | Whitfield, H, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2019-09-04 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Allosteric Site on SHIP2 Identified Through Fluorescent Ligand Screening and Crystallography: A Potential New Target for Intervention.
J.Med.Chem., 64, 2021
|
|
1C5K
 
 | | THE STRUCTURE OF TOLB, AN ESSENTIAL COMPONENT OF THE TOL-DEPENDENT TRANSLOCATION SYSTEM AND ITS INTERACTIONS WITH THE TRANSLOCATION DOMAIN OF COLICIN E9 | | Descriptor: | PROTEIN (TOLB PROTEIN), YTTERBIUM (III) ION | | Authors: | Carr, S, Penfold, C.N, Bamford, V, James, R, Hemmings, A.M. | | Deposit date: | 1999-12-05 | | Release date: | 2000-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structure of TolB, an essential component of the tol-dependent translocation system, and its protein-protein interaction with the translocation domain of colicin E9.
Structure Fold.Des., 8, 2000
|
|
2GZE
 
 | | Crystal structure of the E9 DNase domain with a mutant immunity protein IM9 (Y55A) | | Descriptor: | Colicin-E9, Colicin-E9 immunity protein, PHOSPHATE ION, ... | | Authors: | Santi, P.S, Kolade, O.O, Kuhlmann, U.C, Kleanthous, C, Hemmings, A.M. | | Deposit date: | 2006-05-11 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the Complex of
the Colicin E9 DNase Domain with a Mutant Immunity Protein, IM9 (Y55A)
To be Published
|
|
2GZI
 
 | | Crystal Structure of the E9 DNase Domain with a Mutant Immunity Protein IM9 (V34A) | | Descriptor: | Colicin-E9, Colicin-E9 immunity protein, PHOSPHATE ION, ... | | Authors: | Santi, P.S, Kolade, O.O, Kuhlmann, U.C, Kleanthous, C, Hemmings, A.M. | | Deposit date: | 2006-05-11 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Complex of the Colicin E9 DNase Domain with a Mutant Immunity Protein, IM9 (V34A)
To be Published
|
|
2GZF
 
 | | Crystal structure of the E9 DNase domain with a mutant immunity protein IM9 (Y54F) | | Descriptor: | Colicin-E9, Colicin-E9 immunity protein, PHOSPHATE ION, ... | | Authors: | Santi, P.S, Kolade, O.O, Kuhlmann, U.C, Kleanthous, C, Hemmings, A.M. | | Deposit date: | 2006-05-11 | | Release date: | 2007-07-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal Structure of the Complex of
the Colicin E9 DNase Domain with a Mutant Immunity Protein, IM9 (Y54F)
To be Published
|
|
2GYK
 
 | | Crystal structure of the complex of the Colicin E9 DNase domain with a mutant immunity protein, IMME9 (D51A) | | Descriptor: | Colicin-E9, Colicin-E9 immunity protein, PHOSPHATE ION, ... | | Authors: | Santi, P.S, Kolade, O.O, Kuhlmann, U.C, Hemmings, A.M. | | Deposit date: | 2006-05-09 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of the complexes of the Colicin E9 DNase domain with mutant immunity proteins
To be Published
|
|
2GZJ
 
 | | Crystal Structure of the E9 DNase Domain with a Mutant Immunity Protein IM9 (D51A) | | Descriptor: | Colicin-E9, Colicin-E9 immunity protein, PHOSPHATE ION, ... | | Authors: | Santi, P.S, Kolade, O.O, Kuhlmann, U.C, Kleanthous, C, Hemmings, A.M. | | Deposit date: | 2006-05-11 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Complex of
the Colicin E9 DNase Domain with a Mutant Immunity Protein, IM9 (D51A)
To be Published
|
|
2GZG
 
 | | Crystal Structure of the E9 DNase Domain with a Mutant Immunity Protein IM9 (Y55F) | | Descriptor: | Colicin-E9, Colicin-E9 immunity protein, PHOSPHATE ION, ... | | Authors: | Santi, P.S, Kolade, O.O, Kuhlmann, U.C, Kleanthous, C, Hemmings, A.M. | | Deposit date: | 2006-05-11 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Complex of
the Colicin E9 DNase Domain with a Mutant Immunity Protein, IM9 (Y55F)
To be Published
|
|
6FL8
 
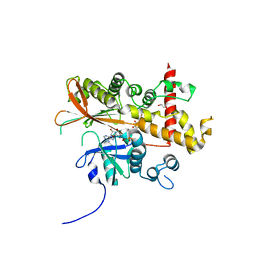 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with purpurogallin and ADP | | Descriptor: | 1,2-ETHANEDIOL, 2,3,4,6-tetrahydroxy-5H-benzo[7]annulen-5-one, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Whitfield, H.L, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-01-25 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Fluorescent Probe Identifies Active Site Ligands of Inositol Pentakisphosphate 2-Kinase.
J. Med. Chem., 61, 2018
|
|
6FJK
 
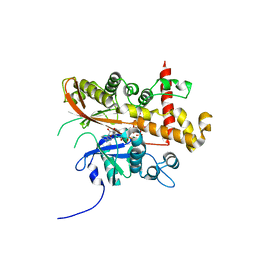 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with myo-IP6 and ADP | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Whitfield, H.L, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-01-22 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.025 Å) | | Cite: | A Fluorescent Probe Identifies Active Site Ligands of Inositol Pentakisphosphate 2-Kinase.
J. Med. Chem., 61, 2018
|
|
6FL3
 
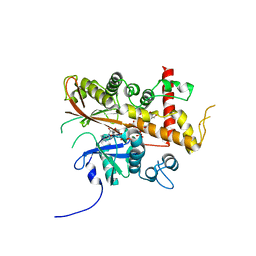 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with myo-IP5 and ADP | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE-5'-DIPHOSPHATE, Inositol-pentakisphosphate 2-kinase, ... | | Authors: | Whitfield, H.L, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-01-25 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | A Fluorescent Probe Identifies Active Site Ligands of Inositol Pentakisphosphate 2-Kinase.
J. Med. Chem., 61, 2018
|
|
6GFG
 
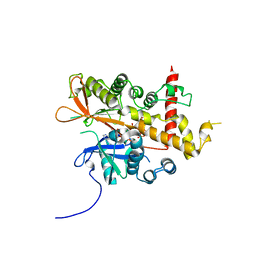 | | Inositol 1,3,4,5,6-pentakisphosphate 2-kinase from A. thaliana in complex with D-chiro-IP6 and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, D-chiro inositol hexakisphosphate, Inositol-pentakisphosphate 2-kinase, ... | | Authors: | Whitfield, H.L, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-04-30 | | Release date: | 2018-09-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Fluorescent Probe Identifies Active Site Ligands of Inositol Pentakisphosphate 2-Kinase.
J. Med. Chem., 61, 2018
|
|
6GIZ
 
 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - SUBSTRATE COMPLEX | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
6GJ2
 
 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - COMPLEX WITH INOSITOL HEXASULPHATE | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
6GIT
 
 | | PURPLE ACID PHYTASE FROM WHEAT ISOFORM B2 - PRODUCT COMPLEX | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Faba-Rodriguez, R, Brearley, C.A, Hemmings, A.M. | | Deposit date: | 2018-05-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.418 Å) | | Cite: | Structure of a cereal purple acid phytase provides new insights to phytate degradation in plants.
Plant Commun., 3, 2022
|
|
