7L6R
 
 | | Crystal Structure of SARS-CoV-2 Nsp16/10 Heterodimer in Complex with (m7GpppA2m)pUpUpApApA (Cap-1), S-Adenosyl-L-homocysteine (SAH) and Manganese (Mn). | | 分子名称: | 2'-O-methyltransferase, CHLORIDE ION, MANGANESE (II) ION, ... | | 著者 | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-12-23 | | 公開日 | 2021-01-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.98 Å) | | 主引用文献 | Mn 2+ coordinates Cap-0-RNA to align substrates for efficient 2'- O -methyl transfer by SARS-CoV-2 nsp16.
Sci.Signal., 14, 2021
|
|
7L6T
 
 | | Crystal Structure of SARS-CoV-2 Nsp16/10 Heterodimer in Complex with (m7GpppA2m)pUpUpApApA (Cap-1), S-Adenosyl-L-homocysteine (SAH) and two Magnesium (Mg) ions. | | 分子名称: | 2'-O-methyltransferase, CHLORIDE ION, FORMIC ACID, ... | | 著者 | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Kiryukhina, O, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-12-23 | | 公開日 | 2021-01-06 | | 最終更新日 | 2024-03-06 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Mn 2+ coordinates Cap-0-RNA to align substrates for efficient 2'- O -methyl transfer by SARS-CoV-2 nsp16.
Sci.Signal., 14, 2021
|
|
6BXG
 
 | | 1.45 Angstrom Resolution Crystal Structure of PDZ domain of Carboxy-Terminal Protease from Vibrio cholerae in Complex with Peptide. | | 分子名称: | CHLORIDE ION, IODIDE ION, LEU-ILE-ALA, ... | | 著者 | Minasov, G, Shuvalova, L, Filippova, E.V, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-12-18 | | 公開日 | 2018-01-03 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | 1.45 Angstrom Resolution Crystal Structure of PDZ domain of Carboxy-Terminal Protease from Vibrio cholerae in Complex with Peptide.
To Be Published
|
|
6BZ0
 
 | | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Acinetobacter baumannii in Complex with FAD. | | 分子名称: | CHLORIDE ION, Dihydrolipoyl dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE, ... | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2017-12-21 | | 公開日 | 2018-01-03 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | 1.83 Angstrom Resolution Crystal Structure of Dihydrolipoyl Dehydrogenase from Acinetobacter baumannii in Complex with FAD.
To Be Published
|
|
6C43
 
 | | 2.9 Angstrom Resolution Crystal Structure of Gamma-Aminobutyraldehyde Dehydrogenase from Salmonella typhimurium. | | 分子名称: | Gamma-aminobutyraldehyde dehydrogenase | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Winsor, J, Tekleab, H, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-01-11 | | 公開日 | 2018-01-24 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | 2.9 Angstrom Resolution Crystal Structure of Gamma-Aminobutyraldehyde Dehydrogenase from Salmonella typhimurium.
To Be Published
|
|
6N1N
 
 | | Crystal structure of class D beta-lactamase from Sebaldella termitidis ATCC 33386 | | 分子名称: | Beta-lactamase, GLYCEROL, SULFATE ION | | 著者 | Michalska, K, Tesar, C, Endres, M, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-11-09 | | 公開日 | 2018-12-19 | | 最終更新日 | 2019-12-18 | | 実験手法 | X-RAY DIFFRACTION (1.601 Å) | | 主引用文献 | Crystal structure of class D beta-lactamase from Sebaldella termitidis ATCC 33386
To Be Published
|
|
6WY4
 
 | | Crystal Structure of Wild Type Class D beta-lactamase from Clostridium difficile 630 | | 分子名称: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SODIUM ION | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-05-12 | | 公開日 | 2020-05-27 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structure of Wild Type Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
7KOS
 
 | | 1.50 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58 | | 分子名称: | FORMIC ACID, MALONIC ACID, Pterin Binding Protein, ... | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Pshenychnyi, S, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-11-09 | | 公開日 | 2021-11-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | 1.50 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58.
To Be Published
|
|
7KOU
 
 | | 1.83 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58 | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Pterin Binding Protein, ... | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Pshenychnyi, S, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-11-10 | | 公開日 | 2021-11-17 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.83 Å) | | 主引用文献 | 1.83 Angstroms Resolution Crystal Structure of Putative Pterin Binding Protein PruR (Atu3496) from Agrobacterium fabrum str. C58.
To Be Published
|
|
7L6J
 
 | | Crystal Structure of the Putative Hydrolase from Stenotrophomonas maltophilia | | 分子名称: | CHLORIDE ION, FORMIC ACID, Putative hydrolase, ... | | 著者 | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-12-23 | | 公開日 | 2021-12-01 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Crystal Structure of the Putative Hydrolase from Stenotrophomonas maltophilia
To Be Published
|
|
7L6Z
 
 | | Crystal Structure of Peptidyl-Prolyl Cis-Trans Isomerasefrom (PpiB) Streptococcus pneumoniae R6 | | 分子名称: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | 著者 | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-12-24 | | 公開日 | 2021-12-01 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Crystal Structure of Peptidyl-Prolyl Cis-Trans Isomerasefrom (PpiB) Streptococcus pneumoniae R6
To Be Published
|
|
7L5T
 
 | | Crystal Structure of the Oxacillin-hydrolyzing Class D Extended-spectrum Beta-lactamase OXA-14 from Pseudomonas aeruginosa in Complex with Covalently Bound Clavulanic Acid | | 分子名称: | (2E)-3-[(4-hydroxy-2-oxobutyl)amino]prop-2-enal, Beta-lactamase, CHLORIDE ION, ... | | 著者 | Minasov, G, Shuvalova, L, Rosas-Lemus, M, Brunzelle, J.S, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID), Center for Membrane Proteins of Infectious Diseases (MPID) | | 登録日 | 2020-12-22 | | 公開日 | 2021-12-29 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Crystal Structure of the Oxacillin-hydrolyzing Class D Extended-spectrum Beta-lactamase OXA-14 from Pseudomonas aeruginosa in Complex with Covalently Bound Clavulanic Acid
To Be Published
|
|
6DB1
 
 | | 2.0 Angstrom Resolution Crystal Structure of N-Terminal Ligand-Binding Domain of Putative Methyl-Accepting Chemotaxis Protein from Salmonella enterica | | 分子名称: | CHLORIDE ION, Putative methyl-accepting chemotaxis protein | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Kiryukhina, O, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-05-02 | | 公開日 | 2018-05-16 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | 2.0 Angstrom Resolution Crystal Structure of N-Terminal Ligand-Binding Domain of Putative Methyl-Accepting Chemotaxis Protein from Salmonella enterica.
To Be Published
|
|
6DLL
 
 | | 2.2 Angstrom Resolution Crystal Structure of P-Hydroxybenzoate Hydroxylase from Pseudomonas putida in Complex with FAD. | | 分子名称: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, FORMIC ACID, ... | | 著者 | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Grimshaw, S, Kwon, K, Anderson, W.F, Satchell, K.J.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2018-06-01 | | 公開日 | 2018-06-13 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Structural comparison of p-hydroxybenzoate hydroxylase (PobA) from Pseudomonas putida with PobA from other Pseudomonas spp. and other monooxygenases.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6XD8
 
 | | Crystal Structure of Peptidylprolyl Isomerase (PrsA) Fragment from Bacillus anthracis | | 分子名称: | Foldase protein PrsA 1 | | 著者 | Minasov, G, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Wiersum, G, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-06-10 | | 公開日 | 2020-07-01 | | 実験手法 | X-RAY DIFFRACTION (1.52 Å) | | 主引用文献 | Crystal Structure of Peptidylprolyl Isomerase (PrsA) Fragment from Bacillus anthracis
To Be Published
|
|
6U7L
 
 | | 2.75 Angstrom Crystal Structure of Galactarate Dehydratase from Escherichia coli. | | 分子名称: | CALCIUM ION, CHLORIDE ION, Galactarate dehydratase (L-threo-forming) | | 著者 | Minasov, G, Shuvalova, L, Wawrzak, Z, Dubrovska, I, Kiryukhina, O, Endres, M, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-09-03 | | 公開日 | 2019-11-06 | | 最終更新日 | 2021-01-27 | | 実験手法 | X-RAY DIFFRACTION (2.75 Å) | | 主引用文献 | Structure of galactarate dehydratase, a new fold in an enolase involved in bacterial fitness after antibiotic treatment.
Protein Sci., 29, 2020
|
|
7RJ1
 
 | | Crystal structure of Aro7p chorismate mutase from Candida albicans, complex with L-Trp | | 分子名称: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Chorismate mutase, ... | | 著者 | Stogios, P.J, Evdokimova, E, Tan, K, Di Leo, R, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-07-20 | | 公開日 | 2021-07-28 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.16 Å) | | 主引用文献 | Crystal structure of Aro7p chorismate mutase from Candida albicans, complex with L-Trp
To Be Published
|
|
7RLR
 
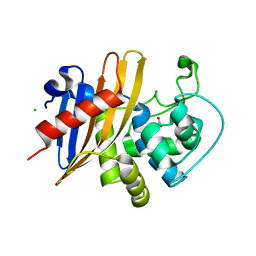 | | Crystal Structure of K83A Mutant of Class D beta-lactamase from Clostridium difficile 630 | | 分子名称: | 1,2-ETHANEDIOL, ACETATE ION, Beta-lactamase, ... | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-07-26 | | 公開日 | 2021-08-11 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (1.88 Å) | | 主引用文献 | Crystal Structure of K83A Mutant of Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
7RL8
 
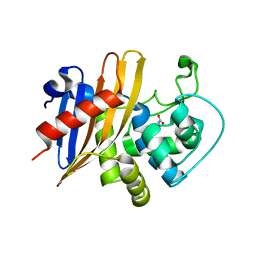 | | Crystal Structure of C79A Mutant of Class D beta-lactamase from Clostridium difficile 630 | | 分子名称: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, SULFATE ION | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2021-07-23 | | 公開日 | 2021-08-11 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal Structure of C79A Mutant of Class D beta-lactamase from Clostridium difficile 630
To Be Published
|
|
3PHY
 
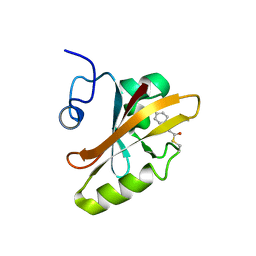 | | PHOTOACTIVE YELLOW PROTEIN, DARK STATE (UNBLEACHED), SOLUTION STRUCTURE, NMR, 26 STRUCTURES | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | 著者 | Dux, P, Rubinstenn, G, Vuister, G.W, Boelens, R, Mulder, F.A.A, Hard, K, Hoff, W.D, Kroon, A, Crielaard, W, Hellingwerf, K.J, Kaptein, R. | | 登録日 | 1998-02-06 | | 公開日 | 1998-05-27 | | 最終更新日 | 2022-03-16 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure and backbone dynamics of the photoactive yellow protein.
Biochemistry, 37, 1998
|
|
6XS4
 
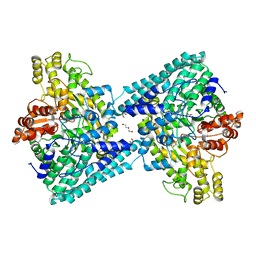 | | Crystal structure of glycyl radical enzyme ECL_02896 from Enterobacter cloacae subsp. cloacae | | 分子名称: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Formate C-acetyltransferase | | 著者 | Valleau, D, Evdokimova, E, Stogios, P.J, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-07-14 | | 公開日 | 2020-08-12 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.33 Å) | | 主引用文献 | Crystal structure of glycyl radical enzyme ECL_02896 from Enterobacter cloacae subsp. cloacae.
To Be Published
|
|
6UE2
 
 | | 1.85 Angstrom Resolution Crystal Structure of Class D beta-lactamase from Clostridium difficile 630 | | 分子名称: | Beta-lactamase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | 著者 | Minasov, G, Shuvalova, L, Dubrovska, I, Rosas-Lemus, M, Jedrzejczak, R, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-09-20 | | 公開日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | 1.85 Angstrom Resolution Crystal Structure of Class D beta-lactamase from Clostridium difficile 630.
To Be Published
|
|
6WON
 
 | | Crystal structure of acetoin dehydrogenase YohF from Salmonella typhimurium | | 分子名称: | CHLORIDE ION, SULFATE ION, YohF | | 著者 | Stogios, P.J, Skarina, T, Mesa, N, Endres, M, Savchenko, A, Joachimiak, A, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2020-04-25 | | 公開日 | 2020-05-13 | | 最終更新日 | 2023-10-18 | | 実験手法 | X-RAY DIFFRACTION (2.13 Å) | | 主引用文献 | Crystal structure of acetoin dehydrogenase YohF from Salmonella typhimurium
To Be Published
|
|
6VBB
 
 | | 2.60 Angstrom Resolution Crystal Structure of Peptidase S41 from Acinetobacter baumannii | | 分子名称: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Peptidase S41, ... | | 著者 | Minasov, G, Wawrzak, Z, Shuvalova, L, Kiryukhina, O, Dubrovska, I, Satchell, K.J.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2019-12-18 | | 公開日 | 2019-12-25 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | 2.60 Angstrom Resolution Crystal Structure of Peptidase S41 from Acinetobacter baumannii
To Be Published
|
|
2PYR
 
 | | PHOTOACTIVE YELLOW PROTEIN, 1 NANOSECOND INTERMEDIATE (287K) | | 分子名称: | 4'-HYDROXYCINNAMIC ACID, PHOTOACTIVE YELLOW PROTEIN | | 著者 | Perman, B, Srajer, V, Ren, Z, Teng, T.Y, Pradervand, C, Ursby, T, Bourgeois, D, Schotte, F, Wulff, M, Kort, R, Hellingwerf, K, Moffat, K. | | 登録日 | 1998-03-04 | | 公開日 | 1999-04-06 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Energy transduction on the nanosecond time scale: early structural events in a xanthopsin photocycle.
Science, 279, 1998
|
|
