6ETT
 
 | | Crystal structure of KDM4D with tetrazole compound 4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.257 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
6ETV
 
 | | Crystal structure of KDM4D with tetrazolhydrazide compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
6ETU
 
 | | Crystal structure of KDM4D with tetrazolhydrazide compound 7 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
6F7D
 
 | |
6ETW
 
 | | Crystal structure of KDM4D with tetrazolhydrazide compound 3 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-27 | | Release date: | 2019-02-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
6F5Q
 
 | | Crystal Structure of KDM4D with GF026 ligand | | Descriptor: | 1,2-ETHANEDIOL, 2-[2-(methylsulfonylamino)ethylamino]pyridine-4-carboxylic acid, CHLORIDE ION, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-12-02 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Crystal Structure of KDM4D with GF026 ligand
To be published
|
|
6F5T
 
 | | Crystal Structure of KDM4D with tetrazole ligand GF057 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-12-02 | | Release date: | 2018-12-12 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal Structure of KDM4D with tetrazole ligand
To be published
|
|
6ETG
 
 | | Crystal structure of KDM4D with tetrazolhydrazide compound 6 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 4D, ... | | Authors: | Malecki, P.H, Link, A, Weiss, M.S, Heinemann, U. | | Deposit date: | 2017-10-26 | | Release date: | 2019-02-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.279 Å) | | Cite: | Structure-Based Screening of Tetrazolylhydrazide Inhibitors versus KDM4 Histone Demethylases.
Chemmedchem, 14, 2019
|
|
6G8Y
 
 | | Crystal Structures of the Single PDZ Domains from GRASP65 and their Interaction with the Golgin GM130 | | Descriptor: | 1,2-ETHANEDIOL, Acetylated cis-Golgi protein, involved in ER-to-Golgi transport, ... | | Authors: | Jurk, C.M, Roske, Y, Heinemann, U. | | Deposit date: | 2018-04-10 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures of the Single PDZ Domains from GRASP65 and their Interaction with the Golgin GM130
Croatica Chemica Acta, 2018
|
|
6G8T
 
 | |
6G8W
 
 | |
5N7H
 
 | |
5N7K
 
 | |
5NXT
 
 | | Wobble base paired RNA double helix | | Descriptor: | RNA (5'-R(*UP*GP*UP*UP*CP*UP*CP*UP*AP*CP*GP*AP*AP*GP*AP*AP*CP*A)-3') | | Authors: | Garg, A, Heinemann, U. | | Deposit date: | 2017-05-10 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.379 Å) | | Cite: | A novel form of RNA double helix based on G·U and C·A+ wobble base pairing.
RNA, 24, 2018
|
|
5RNT
 
 | | X-RAY ANALYSIS OF CUBIC CRYSTALS OF THE COMPLEX FORMED BETWEEN RIBONUCLEASE T1 AND GUANOSINE-3',5'-BISPHOSPHATE | | Descriptor: | GUANOSINE-3',5'-DIPHOSPHATE, RIBONUCLEASE T1 | | Authors: | Saenger, W, Heinemann, U, Lenz, A. | | Deposit date: | 1991-03-28 | | Release date: | 1993-01-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | X-ray analysis of cubic crystals of the complex formed between ribonuclease T1 and guanosine-3',5'-bisphosphate.
Acta Crystallogr.,Sect.B, 47, 1991
|
|
2WAL
 
 | | Crystal Structure of human GADD45gamma | | Descriptor: | GROWTH ARREST AND DNA-DAMAGE-INDUCIBLE PROTEIN GADD45 GAMMA, MALONIC ACID | | Authors: | Bhattacharya, S, Mueller, J.J, Roske, Y, Turnbull, A.P, Quedenau, C, Goetz, F, Buessow, K, Heinemann, U. | | Deposit date: | 2009-02-09 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The Crystal Structure of Human Gadd45Gamma
To be Published
|
|
2W7N
 
 | | Crystal Structure of KorA Bound to Operator DNA: Insight into Repressor Cooperation in RP4 Gene Regulation | | Descriptor: | 5'-D(*AP*AP*TP*GP*TP*TP*TP*AP*GP*CP *TP*AP*AP*AP*CP*AP*AP*G)-3', 5'-D(*CP*BRUP*BRUP*GP*TP*TP*TP*AP*GP*CP*TP*AP *AP*AP*CP*AP*BRUP*T)-3', TRFB TRANSCRIPTIONAL REPRESSOR PROTEIN | | Authors: | Koenig, B, Mueller, J.J, Lanka, E, Heinemann, U. | | Deposit date: | 2008-12-23 | | Release date: | 2009-02-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Kora Bound to Operator DNA: Insight Into Repressor Cooperation in Rp4 Gene Regulation
Nucleic Acids Res., 37, 2009
|
|
2WBU
 
 | | CRYSTAL STRUCTURE OF THE ZINC FINGER DOMAIN OF KLF4 BOUND TO ITS TARGET DNA | | Descriptor: | 5'-D(*DGP*DAP*DGP*DGP*DCP*DGP*DTP* DGP*DGP*DC)-3', 5'-D(*DGP*DCP*DCP*DAP*DCP*DGP*DCP* DCP*DTP*DC)-3', KRUEPPEL-LIKE FACTOR 4, ... | | Authors: | Schuetz, A, Zocher, G, Carstanjen, D, Heinemann, U. | | Deposit date: | 2009-03-05 | | Release date: | 2010-04-07 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Structure of the Klf4 DNA-Binding Domain Links to Self-Renewal and Macrophage Differentiation.
Cell.Mol.Life Sci., 68, 2011
|
|
2WNH
 
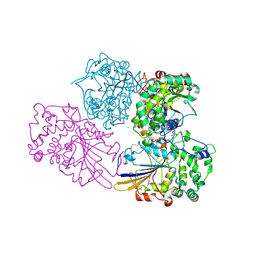 | | Crystal Structure Analysis of Klebsiella sp ASR1 Phytase | | Descriptor: | 3-PHYTASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Bohm, K, Mueller, J.J, Heinemann, U. | | Deposit date: | 2009-07-09 | | Release date: | 2010-04-28 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Crystal Structure of Klebsiella Sp. Asr1 Phytase Suggests Substrate Binding to a Preformed Active Site that Meets the Requirements of a Plant Rhizosphere Enzyme.
FEBS J., 277, 2010
|
|
2WNI
 
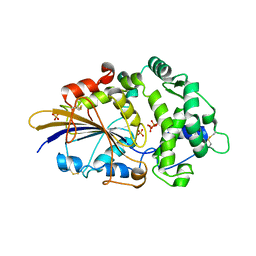 | |
2WU0
 
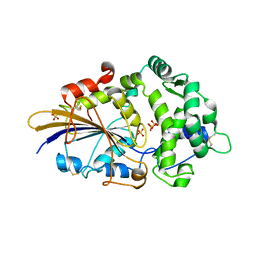 | |
2W3C
 
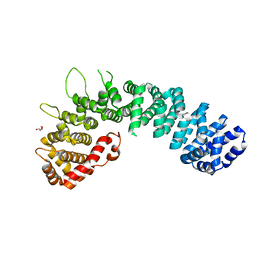 | | Globular head region of the human general vesicular transport factor p115 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GENERAL VESICULAR TRANSPORT FACTOR P115 | | Authors: | Striegl, H, Roske, Y, Kummel, D, Heinemann, U. | | Deposit date: | 2008-11-11 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Unusual Armadillo Fold in the Human General Vesicular Transport Factor P115
Plos One, 4, 2009
|
|
2WBS
 
 | | Crystal structure of the zinc finger domain of Klf4 bound to its target DNA | | Descriptor: | 5'-D(*GP*AP*GP*GP*CP*GP*CP)-3', 5'-D(*GP*CP*GP*CP*CP*TP*CP)-3', GLYCEROL, ... | | Authors: | Zocher, G, Schuetz, A, Carstanjen, D, Heinemann, U. | | Deposit date: | 2009-03-03 | | Release date: | 2010-04-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The Structure of the Klf4 DNA-Binding Domain Links to Self-Renewal and Macrophage Differentiation.
Cell.Mol.Life Sci., 68, 2011
|
|
2VNL
 
 | | MUTANT Y108Wdel OF THE HEADBINDING DOMAIN OF PHAGE P22 TAILSPIKE C- TERMINally fused to ISOLEUCINE ZIPPER pIIGCN4 (chimera II) | | Descriptor: | BIFUNCTIONAL TAIL PROTEIN, PIIGCN4, GLYCEROL | | Authors: | Mueller, J.J, Seul, A, Mueller, G, Seckler, R, Heinemann, U. | | Deposit date: | 2008-02-05 | | Release date: | 2009-02-10 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Bacteriophage P22 Tailspike: Structure of the Complete Protein and Function of the Interdomain Linker
Acta Crystallogr.,Sect.D, 70, 2014
|
|
2WLB
 
 | | Adrenodoxin-like ferredoxin Etp1fd(516-618) of Schizosaccharomyces pombe mitochondria | | Descriptor: | ELECTRON TRANSFER PROTEIN 1, MITOCHONDRIAL, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Mueller, J.J, Hannemann, F, Schiffler, B, Bernhardt, R, Heinemann, U. | | Deposit date: | 2009-06-23 | | Release date: | 2010-08-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural and Thermodynamic Characterization of the Adrenodoxin-Like Domain of the Electron-Transfer Protein Etp1 from Schizosaccharomyces Pombe.
J.Inorg.Biochem., 105, 2011
|
|
