1GLH
 
 | | CATION BINDING TO A BACILLUS (1,3-1,4)-BETA-GLUCANASE. GEOMETRY, AFFINITY AND EFFECT ON PROTEIN STABILITY | | 分子名称: | 1,3-1,4-BETA-GLUCANASE, SODIUM ION | | 著者 | Keitel, T, Heinemann, U. | | 登録日 | 1994-11-25 | | 公開日 | 1995-02-07 | | 最終更新日 | 2024-06-05 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Cation binding to a Bacillus (1,3-1,4)-beta-glucanase. Geometry, affinity and effect on protein stability
Eur.J.Biochem., 222, 1994
|
|
7NDJ
 
 | |
6XYD
 
 | | Crystal structure of Q4D6Q6, a conserved kinetoplastid-specific protein from Trypanosoma cruzi | | 分子名称: | CHLORIDE ION, MAGNESIUM ION, SODIUM ION, ... | | 著者 | Roske, Y, Heinemann, U, Andrea, E.D. | | 登録日 | 2020-01-30 | | 公開日 | 2020-06-10 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.81 Å) | | 主引用文献 | Crystal structure of Q4D6Q6, a conserved kinetoplastid-specific protein from Trypanosoma cruzi.
J.Struct.Biol., 211, 2020
|
|
6Q3V
 
 | |
6TGF
 
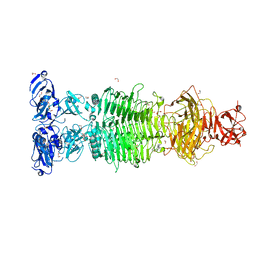 | | Pantoea stewartii WceF is a glycan biofilm modifying enzyme with a bacteriophage tailspike-like parallel beta-helix fold | | 分子名称: | 1,2-ETHANEDIOL, Exopolysaccharide biosynthesis protein, TETRAETHYLENE GLYCOL | | 著者 | Irmscher, T, Roske, Y, Gayk, I, Heinemann, U, Barbirz, S. | | 登録日 | 2019-11-15 | | 公開日 | 2020-11-25 | | 最終更新日 | 2024-05-15 | | 実験手法 | X-RAY DIFFRACTION (2.55 Å) | | 主引用文献 | Pantoea stewartii WceF is a glycan biofilm-modifying enzyme with a bacteriophage tailspike-like fold.
J.Biol.Chem., 296, 2021
|
|
5IFW
 
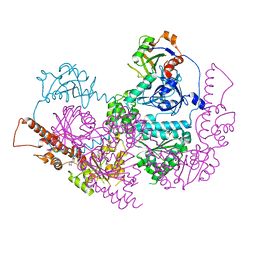 | |
5IFS
 
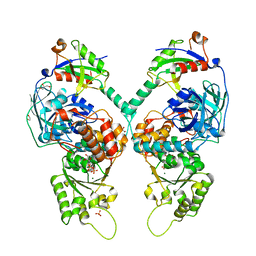 | | Quantitative interaction mapping reveals an extended ubiquitin regulatory domain in ASPL that disrupts functional p97 hexamers and induces cell death | | 分子名称: | 1,2-ETHANEDIOL, ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, ... | | 著者 | Roske, Y, Arumughan, A, Heinemann, U, Wanker, E. | | 登録日 | 2016-02-26 | | 公開日 | 2016-10-26 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.46 Å) | | 主引用文献 | Quantitative interaction mapping reveals an extended UBX domain in ASPL that disrupts functional p97 hexamers.
Nat Commun, 7, 2016
|
|
1MJC
 
 | |
4XLE
 
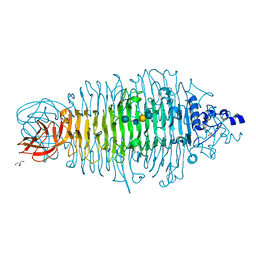 | | Tailspike protein double mutant D339N/E372A of E. coli bacteriophage HK620 in complex with hexasaccharide | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | 著者 | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | 登録日 | 2015-01-13 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
4XKW
 
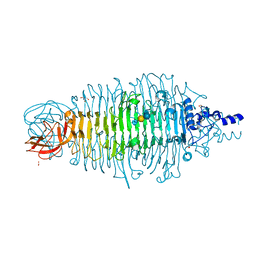 | | Tailspike protein mutant D339N of E. coli bacteriophage HK620 in complex with pentasaccharide | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | 著者 | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | 登録日 | 2015-01-12 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.45 Å) | | 主引用文献 | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
4XLA
 
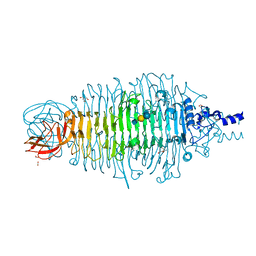 | | Tailspike protein mutant D339A of E. coli bacteriophage HK620 IN COMPLEX WITH PENTASACCHARIDE | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | 著者 | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | 登録日 | 2015-01-13 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
4XM3
 
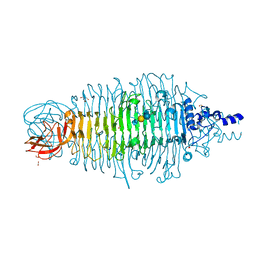 | | Tailspike protein mutant E372A of E. coli bacteriophage HK620 in complex with pentasaccharide | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | 著者 | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | 登録日 | 2015-01-14 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.27 Å) | | 主引用文献 | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
4XN3
 
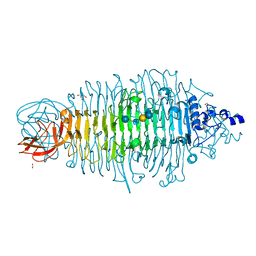 | | Tailspike protein mutant E372A of E. coli bacteriophage HK620 in complex with hexasaccharide | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | 著者 | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | 登録日 | 2015-01-15 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
4XON
 
 | | Tailspike protein double mutant D339N/E372Q of E. coli bacteriophage HK620 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | 著者 | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | 登録日 | 2015-01-16 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
4XR6
 
 | | Tailspike protein double mutant D339A/E372Q of E. coli bacteriophage HK620 in complex with hexasaccharide | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | 著者 | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | 登録日 | 2015-01-20 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.78 Å) | | 主引用文献 | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
4XOT
 
 | | Tailspike protein mutant E372Q of E. coli bacteriophage HK620 in complex with pentasaccharide | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | 著者 | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | 登録日 | 2015-01-16 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.06 Å) | | 主引用文献 | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
4XLH
 
 | | Tailspike protein double mutant D339A/E372A of E. coli bacteriophage HK620 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | 著者 | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | 登録日 | 2015-01-13 | | 公開日 | 2016-01-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.91 Å) | | 主引用文献 | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
4XQF
 
 | | Tailspike protein mutant E372Q (delta D470/N471) of E. coli bacteriophage HK620 | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | 著者 | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | 登録日 | 2015-01-19 | | 公開日 | 2016-01-27 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
4YEJ
 
 | | Tailspike protein double mutant D339A/E372Q of E. coli bacteriophage HK620 in complex with pentasaccharide | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | 著者 | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | 登録日 | 2015-02-24 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.4 Å) | | 主引用文献 | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
4YEL
 
 | | Tailspike protein double mutant D339A/E372A of E. coli bacteriophage HK620 in complex with hexasaccharide | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, SODIUM ION, ... | | 著者 | Gohlke, U, Broeker, N.K, Heinemann, U, Seckler, R, Barbirz, S. | | 登録日 | 2015-02-24 | | 公開日 | 2016-03-09 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.72 Å) | | 主引用文献 | Enthalpic cost of water removal from a hydrophobic glucose binding cavity on HK620 tailspike protein.
to be published
|
|
7ZIJ
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-05-080 | | 分子名称: | 8-(1~{H}-benzimidazol-2-ylmethyl)-3-cyclopropyl-7-(phenylmethyl)purine-2,6-dione, FE (III) ION, Tryptophan 5-hydroxylase 1 | | 著者 | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | 登録日 | 2022-04-08 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.94678366 Å) | | 主引用文献 | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZIF
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-480 | | 分子名称: | (2R)-2-azanyl-5-[[2-[3-methyl-2,6-bis(oxidanylidene)-7-(phenylmethyl)purin-8-yl]sulfanyl-3H-benzimidazol-5-yl]amino]-5-oxidanylidene-pentanoic acid, FE (III) ION, GLYCEROL, ... | | 著者 | Schuetz, A, Ziebart, N, Weise, M, Mallow, K, Pfeifer, J, Nazare, M, Specker, E, Heinemann, U. | | 登録日 | 2022-04-08 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.86859715 Å) | | 主引用文献 | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZIH
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor AG-01-128 | | 分子名称: | 8-(1~{H}-benzimidazol-2-ylmethyl)-3-ethyl-7-(phenylmethyl)purine-2,6-dione, DI(HYDROXYETHYL)ETHER, FE (III) ION, ... | | 著者 | Schuetz, A, Gogolin, A, Pfeifer, J, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | 登録日 | 2022-04-08 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.46890831 Å) | | 主引用文献 | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZII
 
 | | Crystal structure of human tryptophan hydroxylase 1 in complex with inhibitor KM-05-193 | | 分子名称: | 8-(5~{H}-[1,3]dioxolo[4,5-f]benzimidazol-6-ylmethyl)-7-(phenylmethyl)-3-propyl-purine-2,6-dione, FE (III) ION, GLYCEROL, ... | | 著者 | Schuetz, A, Mallow, K, Nazare, M, Specker, E, Heinemann, U. | | 登録日 | 2022-04-08 | | 公開日 | 2022-09-07 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.6280005 Å) | | 主引用文献 | Structure-Based Design of Xanthine-Benzimidazole Derivatives as Novel and Potent Tryptophan Hydroxylase Inhibitors.
J.Med.Chem., 65, 2022
|
|
7ZIK
 
 | |
