5EI0
 
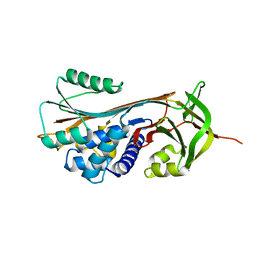 | | Structure of RCL-cleaved vaspin (serpinA12) | | Descriptor: | Serpin A12 | | Authors: | Pippel, J, Kuettner, B.E, Ulbricht, D, Daberger, J, Schultz, S, Heiker, J.T, Strater, N. | | Deposit date: | 2015-10-29 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of cleaved vaspin (serpinA12).
Biol.Chem., 397, 2016
|
|
4Y3K
 
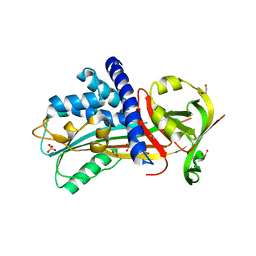 | | Structure of Vaspin mutant E379S | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Serpin A12 | | Authors: | Pippel, J, Strater, N, Ulbricht, D, Schultz, S, Meier, R, Heiker, J.T. | | Deposit date: | 2015-02-10 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A unique serpin P1' glutamate and a conserved beta-sheet C arginine are key residues for activity, protease recognition and stability of serpinA12 (vaspin).
Biochem.J., 470, 2015
|
|
4Y40
 
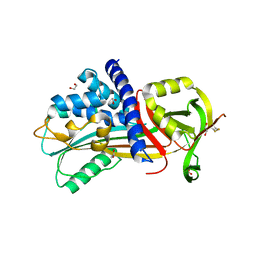 | | Structure of Vaspin mutant D305C V383C | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, Serpin A12 | | Authors: | Pippel, J, Strater, N, Ulbricht, D, Schultz, S, Meier, R, Heiker, J.T. | | Deposit date: | 2015-02-10 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A unique serpin P1' glutamate and a conserved beta-sheet C arginine are key residues for activity, protease recognition and stability of serpinA12 (vaspin).
Biochem.J., 470, 2015
|
|
4IF8
 
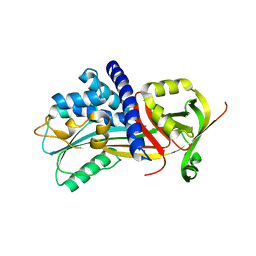 | | Structure Of Vaspin | | Descriptor: | SULFATE ION, Serpin A12 | | Authors: | Kuettner, E.B, Strater, N, Heiker, J.T, Kloting, N, Kovacs, P, Schultz, S, Kern, M, Stumvoll, M, Bluher, M, Beck-Sickinger, A.G. | | Deposit date: | 2012-12-14 | | Release date: | 2013-02-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Vaspin inhibits kallikrein 7 by serpin mechanism
Cell.Mol.Life Sci., 70, 2013
|
|
