3HWP
 
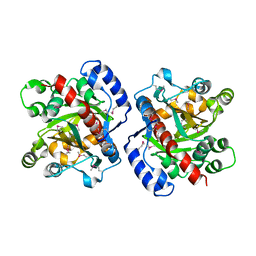 | | Crystal structure and computational analyses provide insights into the catalytic mechanism of 2, 4-diacetylphloroglucinol hydrolase PhlG from Pseudomonas fluorescens | | Descriptor: | CHLORIDE ION, NICKEL (II) ION, PhlG, ... | | Authors: | He, Y.-X, Huang, L, Xue, Y, Fei, X, Teng, Y.-B, Zhou, C.-Z. | | Deposit date: | 2009-06-18 | | Release date: | 2009-12-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure and Computational Analyses Provide Insights into the Catalytic Mechanism of 2,4-Diacetylphloroglucinol Hydrolase PhlG from Pseudomonas fluorescens.
J.Biol.Chem., 285, 2010
|
|
8ZMD
 
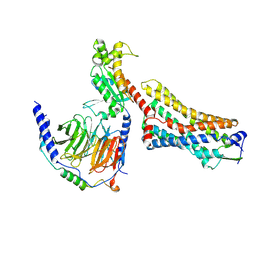 | | Protease-activated receptor-2 (PAR2)/Gq complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Proteinase-activated receptor 2, ... | | Authors: | He, Y, Zhu, X. | | Deposit date: | 2024-05-23 | | Release date: | 2025-05-28 | | Last modified: | 2025-12-10 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis of protease-activated receptor 2 activation and biased agonism.
Cell Discov, 11, 2025
|
|
8ZME
 
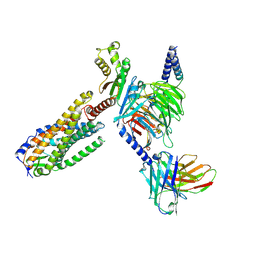 | | Protease-activated receptor-2 (PAR2)/miniG13 complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Proteinase-activated receptor 2, ... | | Authors: | He, Y, Zhu, X. | | Deposit date: | 2024-05-23 | | Release date: | 2025-05-28 | | Last modified: | 2025-12-10 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis of protease-activated receptor 2 activation and biased agonism.
Cell Discov, 11, 2025
|
|
1ZXH
 
 | | G311 mutant protein | | Descriptor: | Immunoglobulin G binding protein G | | Authors: | He, Y, Yeh, D.C, Alexander, P, Bryan, P.N, Orban, J. | | Deposit date: | 2005-06-08 | | Release date: | 2005-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structures of IgG binding domains with artificially evolved high levels of sequence identity but different folds.
Biochemistry, 44, 2005
|
|
1ZXG
 
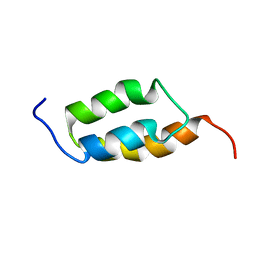 | | Solution structure of A219 | | Descriptor: | Immunoglobulin G binding protein A | | Authors: | He, Y, Yeh, D.C, Alexander, P, Bryan, P.N, Orban, J. | | Deposit date: | 2005-06-08 | | Release date: | 2005-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structures of IgG binding domains with artificially evolved high levels of sequence identity but different folds.
Biochemistry, 44, 2005
|
|
9UET
 
 | | Cryo-EM structure of human choline-phosphotransferase 1 | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Cholinephosphotransferase 1, MAGNESIUM ION | | Authors: | He, Y.L, Qian, H.W. | | Deposit date: | 2025-04-09 | | Release date: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Cryo-EM structure of human choline-phosphotransferase 1
To Be Published
|
|
4AIU
 
 | | A complex structure of BtGH84 | | Descriptor: | (3AR,5R,6S,7R,7AR)-2,5-BIS(HYDROXYMETHYL)-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]OXAZOLE-6,7-DIOL, CALCIUM ION, O-GLCNACASE BT_4395 | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2012-02-13 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Metabolism of Vertebrate Amino Sugars with N-Glycolyl Groups: Intracellular Beta-O-Linked N-Glycolylglucosamine (Glcngc), Udp-Glcngc, and the Biochemical and Structural Rationale for the Substrate Tolerance of Beta-O-Linked Beta-N-Acetylglucosaminidase.
J.Biol.Chem., 287, 2012
|
|
4AIS
 
 | | A complex structure of BtGH84 | | Descriptor: | GLYCEROL, GLYCOLIC ACID, O-GLCNACASE BT_4395 | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2012-02-13 | | Release date: | 2012-06-20 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metabolism of Vertebrate Amino Sugars with N-Glycolyl Groups: Intracellular Beta-O-Linked N-Glycolylglucosamine (Glcngc), Udp-Glcngc, and the Biochemical and Structural Rationale for the Substrate Tolerance of Beta-O-Linked Beta-N-Acetylglucosaminidase.
J.Biol.Chem., 287, 2012
|
|
7VKT
 
 | | cryo-EM structure of LTB4-bound BLT1 in complex with Gi protein | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Wang, N. | | Deposit date: | 2021-10-01 | | Release date: | 2022-03-09 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structural basis of leukotriene B4 receptor 1 activation.
Nat Commun, 13, 2022
|
|
4BMH
 
 | | Crystal structure of SsHAT | | Descriptor: | ACETYLTRANSFERASE, CHLORIDE ION | | Authors: | He, Y, Turkenburg, J.P, Davies, G.J. | | Deposit date: | 2013-05-08 | | Release date: | 2014-01-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Three-Dimensional Structure of a Streptomyces Sviceus Gnat Acetyltransferase with Similarity to the C-Terminal Domain of the Human Gh84 O-Glcnacase
Acta Crystallogr.,Sect.D, 70, 2014
|
|
8KGG
 
 | | LPS-bound P2Y10 in complex with G13 | | Descriptor: | (2~{S})-2-$l^{4}-azanyl-3-[[(2~{R})-3-octadecanoyloxy-2-oxidanyl-propoxy]-oxidanyl-oxidanylidene-$l^{6}-phosphanyl]oxy-propanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Yin, Y. | | Deposit date: | 2023-08-19 | | Release date: | 2024-07-31 | | Last modified: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Insights into lysophosphatidylserine recognition and G alpha 12/13 -coupling specificity of P2Y10.
Cell Chem Biol, 31, 2024
|
|
9IXJ
 
 | | histamine-bound H2R in complex with Gs | | Descriptor: | CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Chen, K. | | Deposit date: | 2024-07-28 | | Release date: | 2025-07-30 | | Method: | ELECTRON MICROSCOPY (2.92 Å) | | Cite: | histamine-bound H2R in complex with Gs
To Be Published
|
|
7YDJ
 
 | | Cryo EM structure of CD97/miniG12 complex | | Descriptor: | Adhesion G protein-coupled receptor E5 subunit beta, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Wang, N. | | Deposit date: | 2022-07-04 | | Release date: | 2023-07-12 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Cryo EM structure of CD97/miniG12 complex
To Be Published
|
|
2APN
 
 | |
6PO2
 
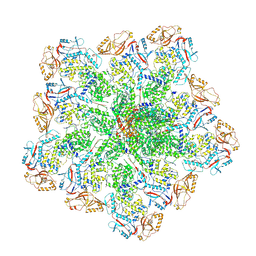 | | In situ structure of BTV RNA-dependent RNA polymerase in BTV core | | Descriptor: | Inner core structural protein VP3, RNA-directed RNA polymerase | | Authors: | He, Y, Shivakoti, S, Ding, K, Cui, Y, Roy, P, Zhou, Z.H. | | Deposit date: | 2019-07-03 | | Release date: | 2019-08-07 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | In situ structures of RNA-dependent RNA polymerase inside bluetongue virus before and after uncoating.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6PNS
 
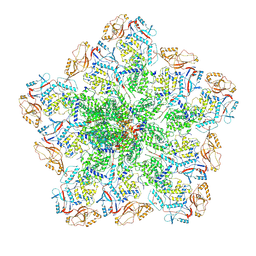 | | In situ structure of BTV RNA-dependent RNA polymerase in BTV virion | | Descriptor: | Inner core structural protein VP3, RNA-directed RNA polymerase | | Authors: | He, Y, Shivakoti, S, Ding, K, Cui, Y, Roy, P, Zhou, Z.H. | | Deposit date: | 2019-07-03 | | Release date: | 2019-08-07 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | In situ structures of RNA-dependent RNA polymerase inside bluetongue virus before and after uncoating.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2XM2
 
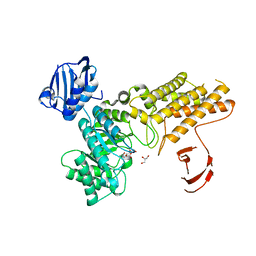 | | BtGH84 in complex with LOGNAc | | Descriptor: | GLYCEROL, N-acetylglucosaminono-1,5-lactone (Z)-oxime, O-GLCNACASE BT_4395 | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2010-07-22 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Inhibition of a Bacterial O-Glcnacase Homologue by Lactone and Lactam Derivatives: Structural, Kinetic and Thermodynamic Analyses.
Amino Acids, 40, 2011
|
|
8ZX4
 
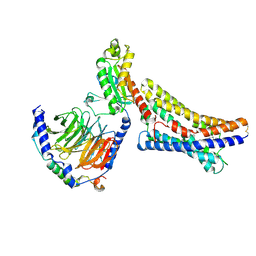 | | LPI-bound GPR55 in complex with G13 | | Descriptor: | G-protein coupled receptor 55, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | He, Y, Xia, R. | | Deposit date: | 2024-06-13 | | Release date: | 2024-11-13 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Structural insight into GPR55 ligand recognition and G-protein coupling.
Cell Res., 35, 2025
|
|
2WCA
 
 | | BtGH84 in complex with n-butyl pugnac | | Descriptor: | CALCIUM ION, O-GLCNACASE BT_4395, [[(3R,4R,5S,6R)-3-(BUTANOYLAMINO)-4,5-DIHYDROXY-6-(HYDROXYMETHYL)OXAN-2-YLIDENE]AMINO] N-PHENYLCARBAMATE | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2009-03-10 | | Release date: | 2009-06-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Insight Into a Strategy for Attenuating Ampc- Mediated Beta-Lactam Resistance: Structural Basis for Selective Inhibition of the Glycoside Hydrolase Nagz.
Protein Sci., 18, 2009
|
|
2X0H
 
 | | BtGH84 Michaelis complex | | Descriptor: | 3,4-difluorophenyl 2-deoxy-2-[(difluoroacetyl)amino]-beta-D-glucopyranoside, CALCIUM ION, GLYCEROL, ... | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2009-12-08 | | Release date: | 2010-01-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Visualizing the Reaction Coordinate of an O-Glcnac Hydrolase
J.Am.Chem.Soc., 132, 2010
|
|
5I8Q
 
 | | S. cerevisiae Prp43 in complex with RNA and ADPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Pre-mRNA-splicing factor ATP-dependent RNA helicase PRP43, ... | | Authors: | He, Y, Nielsen, K.H, Andersen, G.R. | | Deposit date: | 2016-02-19 | | Release date: | 2017-02-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structure of the DEAH/RHA ATPase Prp43p bound to RNA implicates a pair of hairpins and motif Va in translocation along RNA.
RNA, 23, 2017
|
|
8ZX5
 
 | | AM251-bound GPR55 in complex with G13 | | Descriptor: | 1-(2,4-dichlorophenyl)-5-(4-iodophenyl)-4-methyl-~{N}-piperidin-1-yl-pyrazole-3-carboxamide, G-protein coupled receptor 55, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Xia, R. | | Deposit date: | 2024-06-13 | | Release date: | 2024-11-13 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insight into GPR55 ligand recognition and G-protein coupling.
Cell Res., 35, 2025
|
|
9ITB
 
 | | LPA-bound LPAR6 in complex with miniGq | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2024-07-19 | | Release date: | 2025-01-15 | | Last modified: | 2025-07-23 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Molecular mechanism of ligand recognition and activation of lysophosphatidic acid receptor LPAR6.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
9ITE
 
 | | LPA-bound LPAR6 in complex with miniG13 | | Descriptor: | (2R)-2-hydroxy-3-(phosphonooxy)propyl (9E)-octadec-9-enoate, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | He, Y, Duan, Y. | | Deposit date: | 2024-07-19 | | Release date: | 2025-01-15 | | Last modified: | 2025-02-05 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Molecular mechanism of ligand recognition and activation of lysophosphatidic acid receptor LPAR6.
Proc.Natl.Acad.Sci.USA, 122, 2025
|
|
2VVN
 
 | | BtGH84 in complex with NH-Butylthiazoline | | Descriptor: | (3AR,5R,6S,7R,7AR)-2-(ETHYLAMINO)-5-(HYDROXYMETHYL)-5,6,7,7A-TETRAHYDRO-3AH-PYRANO[3,2-D][1,3]THIAZOLE-6,7-DIOL, AMMONIUM ION, GLYCEROL, ... | | Authors: | He, Y, Davies, G.J. | | Deposit date: | 2008-06-10 | | Release date: | 2008-07-01 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Potent Mechanism-Inspired O-Glcnacase Inhibitor that Blocks Phosphorylation of Tau in Vivo.
Nat.Chem.Biol., 4, 2008
|
|
