5H4S
 
 | |
6M5M
 
 | | SPL-1 - GlcNAc complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, N-acetylglucosamine-specific lectin | | Authors: | Unno, H, Hatakeyama, T. | | Deposit date: | 2020-03-11 | | Release date: | 2020-06-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Novel carbohydrate-recognition mode of the invertebrate C-type lectin SPL-1 from Saxidomus purpuratus revealed by the GlcNAc-complex crystal in the presence of Ca2.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
2ZWB
 
 | | Neutron crystal structure of wild type human lysozyme in D2O | | Descriptor: | Lysozyme C | | Authors: | Chiba-Kamoshida, K, Matsui, T, Chatake, T, Ohhara, T, Ostermann, A, Tanaka, I, Yutani, K, Niimura, N. | | Deposit date: | 2008-12-02 | | Release date: | 2009-12-08 | | Last modified: | 2024-10-16 | | Method: | NEUTRON DIFFRACTION (1.8 Å) | | Cite: | Site-specific softening of peptide bonds by localized deuterium observed by neutron crystallography of human lysozyme
To be Published
|
|
3AU4
 
 | |
1I3T
 
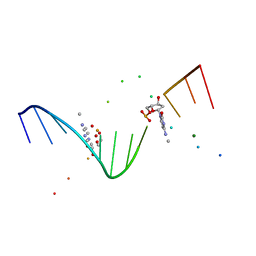 | | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGAATT(MO4)CGCG): THE WATSON-CRICK TYPE AND WOBBLE N4-METHOXYCYTIDINE/GUANOSINE BASE PAIRS IN B-DNA | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(C45)P*GP*CP*GP)-3', MAGNESIUM ION | | Authors: | Hossain, M.T, Hikima, T, Chatake, T, Masaru, T, Sunami, T, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2001-02-16 | | Release date: | 2001-09-14 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic studies on damaged DNAs: III. N(4)-methoxycytosine can form both Watson-Crick type and wobbled base pairs in a B-form duplex.
J.Biochem.(Tokyo), 130, 2001
|
|
1J8L
 
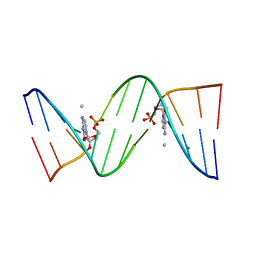 | | Molecular and Crystal Structure of D(CGCAAATTMO4CGCG): the Watson-Crick Type N4-Methoxycytidine/Adenosine Base Pair in B-DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*(C45)P*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Hossain, M.T, Sunami, T, Tsunoda, M, Hikima, T, Chatake, T, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2001-05-22 | | Release date: | 2001-09-28 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic studies on damaged DNAs IV. N(4)-methoxycytosine shows a second face for Watson-Crick base-pairing, leading to purine transition mutagenesis.
Nucleic Acids Res., 29, 2001
|
|
1I47
 
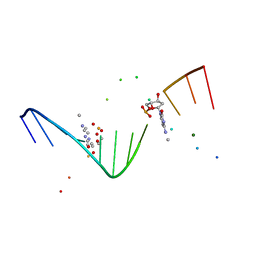 | | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGAATT(MO4)CGCG): THE WATSON-CRICK TYPE AND WOBBLE N4-METHOXYCYTIDINE/GUANOSINE BASE PAIRS IN B-DNA | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*(C45)P*GP*CP*GP)-3', MAGNESIUM ION | | Authors: | Hossain, M.T, Hikima, T, Chatake, T, Tsunoda, M, Sunami, T, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2001-02-20 | | Release date: | 2003-02-11 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic studies on damaged DNAs: III. N(4)-methoxycytosine can form both Watson-Crick type and wobbled base pairs in a B-form duplex
J.Biochem.(Tokyo), 130, 2001
|
|
4Y7E
 
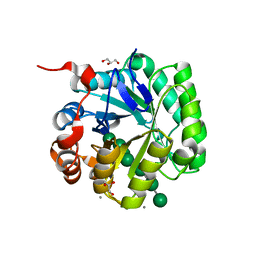 | | Crystal structure of beta-mannanase from Streptomyces thermolilacinus with mannohexaose | | Descriptor: | CALCIUM ION, Endoglucanase, GLYCEROL, ... | | Authors: | Kumagai, Y, Yamashita, K, Okuyama, M, Hatanaka, T, Yao, M, Kimura, A. | | Deposit date: | 2015-02-14 | | Release date: | 2015-09-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The loop structure of Actinomycete glycoside hydrolase family 5 mannanases governs substrate recognition
Febs J., 282, 2015
|
|
7DEQ
 
 | | Lysozyme-sugar complex in D2O | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Lysozyme C, ... | | Authors: | Tanaka, I, Chatake, T. | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Recent structural insights into the mechanism of lysozyme hydrolysis.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
7DER
 
 | | Lysozyme alone in H2O | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION | | Authors: | Tanaka, I, Chatake, T. | | Deposit date: | 2020-11-04 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Recent structural insights into the mechanism of lysozyme hydrolysis.
Acta Crystallogr D Struct Biol, 77, 2021
|
|
1VCL
 
 | | Crystal Structure of Hemolytic Lectin CEL-III | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Uchida, T, Yamasaki, T, Eto, S, Sugawara, H, Kurisu, G, Nakagawa, A, Kusunoki, M, Hatakeyama, T. | | Deposit date: | 2004-03-09 | | Release date: | 2004-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the Hemolytic Lectin CEL-III Isolated from the Marine Invertebrate Cucumaria echinata: IMPLICATIONS OF DOMAIN STRUCTURE FOR ITS MEMBRANE PORE-FORMATION MECHANISM
J.Biol.Chem., 279, 2004
|
|
1WMZ
 
 | | Crystal Structure of C-type Lectin CEL-I complexed with N-acetyl-D-galactosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Sugawara, H, Kusunoki, M, Kurisu, G, Fujimoto, T, Aoyagi, H, Hatakeyama, T. | | Deposit date: | 2004-07-22 | | Release date: | 2004-09-07 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Characteristic Recognition of N-Acetylgalactosamine by an Invertebrate C-type Lectin, CEL-I, Revealed by X-ray Crystallographic Analysis
J.Biol.Chem., 279, 2004
|
|
1WMY
 
 | | Crystal Structure of C-type Lectin CEL-I from Cucumaria echinata | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, lectin CEL-I, ... | | Authors: | Sugawara, H, Kusunoki, M, Kurisu, G, Fujimoto, T, Aoyagi, H, Hatakeyama, T. | | Deposit date: | 2004-07-22 | | Release date: | 2004-09-07 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Characteristic Recognition of N-Acetylgalactosamine by an Invertebrate C-type Lectin, CEL-I, Revealed by X-ray Crystallographic Analysis
J.Biol.Chem., 279, 2004
|
|
3WSU
 
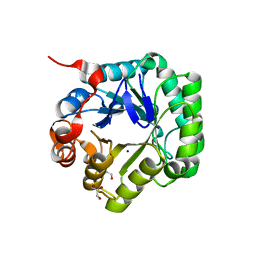 | | Crystal structure of beta-mannanase from Streptomyces thermolilacinus | | Descriptor: | Beta-mannanase, GLYCEROL, SODIUM ION | | Authors: | Kumagai, Y, Yamashita, K, Okuyama, M, Hatanaka, T, Yao, M, Kimura, A. | | Deposit date: | 2014-03-26 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The loop structure of Actinomycete glycoside hydrolase family 5 mannanases governs substrate recognition
Febs J., 282, 2015
|
|
1WN0
 
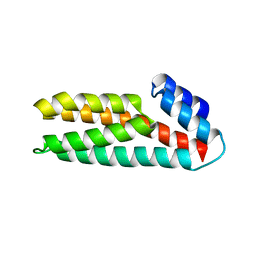 | | Crystal Structure of Histidine-containing Phosphotransfer Protein, ZmHP2, from maize | | Descriptor: | histidine-containing phosphotransfer protein | | Authors: | Sugawara, H, Kawano, Y, Hatakeyama, T, Yamaya, T, Kamiya, N, Sakakibara, H, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-07-24 | | Release date: | 2005-01-25 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the histidine-containing phosphotransfer protein ZmHP2 from maize
Protein Sci., 14, 2005
|
|
1WQZ
 
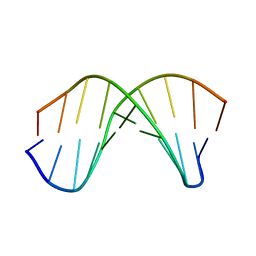 | | Complicated water orientations in the minor groove of B-DNA decamer D(CCATTAATGG)2 observed by neutron diffraction measurements | | Descriptor: | 5'-D(*CP*CP*AP*TP*TP*AP*AP*TP*GP*G)-3' | | Authors: | Arai, S, Chatake, T, Ohhara, T, Kurihara, K, Tanaka, I, Suzuki, N, Fujimoto, Z, Mizuno, H, Niimura, N. | | Deposit date: | 2004-10-07 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (3 Å) | | Cite: | Complicated water orientations in the minor groove of the B-DNA decamer d(CCATTAATGG)2 observed by neutron diffraction measurements
Nucleic Acids Res., 33, 2005
|
|
1WQY
 
 | | X-RAY structural analysis of B-DNA decamer D(CCATTAATGG)2 crystal grown in D2O solution | | Descriptor: | 5'-D(*CP*CP*AP*TP*TP*AP*AP*TP*GP*G)-3' | | Authors: | Arai, S, Chatake, T, Ohhara, T, Kurihara, K, Tanaka, I, Suzuki, N, Fujimoto, Z, Mizuno, H, Niimura, N. | | Deposit date: | 2004-10-07 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Complicated water orientations in the minor groove of the B-DNA decamer d(CCATTAATGG)2 observed by neutron diffraction measurements
Nucleic Acids Res., 33, 2005
|
|
6A7S
 
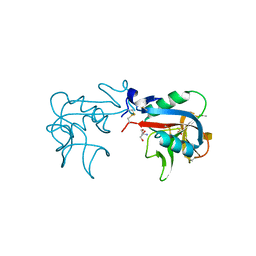 | | Ca2+-independent C-type lectin SPL-2 from Saxidomus purpuratus | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Unno, H, Hatakeyama, T. | | Deposit date: | 2018-07-04 | | Release date: | 2019-03-20 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel Ca2+-independent carbohydrate recognition of the C-type lectins, SPL-1 and SPL-2, from the bivalve Saxidomus purpuratus.
Protein Sci., 28, 2019
|
|
6A56
 
 | | AJLec from the Sea Anemone Anthopleura japonica | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, AJLec, CALCIUM ION, ... | | Authors: | Unno, H, Hatakeyama, T. | | Deposit date: | 2018-06-22 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Identification, Characterization, and X-ray Crystallographic Analysis of a Novel Type of Lectin AJLec from the Sea Anemone Anthopleura japonica.
Sci Rep, 8, 2018
|
|
6A7T
 
 | |
3W9T
 
 | | pore-forming CEL-III | | Descriptor: | CALCIUM ION, Hemolytic lectin CEL-III, MAGNESIUM ION, ... | | Authors: | Unno, H, Goda, S, Hatakeyama, T. | | Deposit date: | 2013-04-16 | | Release date: | 2014-03-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Hemolytic lectin CEL-III heptamer reveals its transmembrane pore-formation mechanism
J.Biol.Chem., 2014
|
|
1VCX
 
 | | Neutron Crystal Structure of the Wild Type Rubredoxin from Pyrococcus Furiosus at 1.5A Resolution | | Descriptor: | FE (III) ION, Rubredoxin | | Authors: | Kurihara, K, Tanaka, I, Chatake, T, Adams, M.W.W, Jenney Jr, F.E, Moiseeva, N, Bau, R, Niimura, N. | | Deposit date: | 2004-03-17 | | Release date: | 2004-08-10 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (1.5 Å) | | Cite: | Neutron crystallographic study on rubredoxin from Pyrococcus furiosus by BIX-3, a single-crystal diffractometer for biomacromolecules
Proc.Natl.Acad.Sci.USA, 101, 2004
|
|
