1UAX
 
 | |
4DWW
 
 | |
3ALU
 
 | | Crystal structure of CEL-IV complexed with Raffinose | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Lectin CEL-IV, ... | | Authors: | Hatakeyama, T, Hozawa, T, Ishii, K, Kamiya, T, Goda, S, Kusunoki, M, Unno, H. | | Deposit date: | 2010-08-07 | | Release date: | 2011-01-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Galactose recognition by a tetrameric C-type lectin, CEL-IV, containing the EPN carbohydrate recognition motif
J.Biol.Chem., 286, 2011
|
|
3ALT
 
 | | Crystal structure of CEL-IV complexed with Melibiose | | Descriptor: | CALCIUM ION, Lectin CEL-IV, C-type, ... | | Authors: | Hatakeyama, T, Hozawa, T, Ishii, K, Kamiya, T, Goda, S, Kusunoki, M, Unno, H. | | Deposit date: | 2010-08-07 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Galactose recognition by a tetrameric C-type lectin, CEL-IV, containing the EPN carbohydrate recognition motif
J.Biol.Chem., 286, 2011
|
|
3ALS
 
 | | Crystal structure of CEL-IV | | Descriptor: | CALCIUM ION, Lectin CEL-IV, C-type | | Authors: | Hatakeyama, T, Hozawa, T, Ishii, K, Kamiya, T, Goda, S, Kusunoki, M, Unno, H. | | Deposit date: | 2010-08-07 | | Release date: | 2011-01-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Galactose recognition by a tetrameric C-type lectin, CEL-IV, containing the EPN carbohydrate recognition motif
J.Biol.Chem., 286, 2011
|
|
8CQ3
 
 | |
8CQ4
 
 | |
8CQ6
 
 | |
8PNJ
 
 | | Chorismate mutase | | Descriptor: | 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Khatanbaatar, T, Cordara, G, Krengel, U. | | Deposit date: | 2023-06-30 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.355 Å) | | Cite: | Structural analysis of chorismate mutase and cyclohexadienyl dehydratase from Pseudomonas aeruginosa
To Be Published
|
|
8PNH
 
 | | Chorismate mutase | | Descriptor: | 3-PHENYLPYRUVIC ACID, 8-HYDROXY-2-OXA-BICYCLO[3.3.1]NON-6-ENE-3,5-DICARBOXYLIC ACID, Bifunctional cyclohexadienyl dehydratase/chorismate mutase from Janthinobacterium sp. HH01, ... | | Authors: | Khatanbaatar, T, Cordara, G, Krengel, U. | | Deposit date: | 2023-06-30 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Chorismate mutase
To Be Published
|
|
8PNI
 
 | | Chorismate mutase | | Descriptor: | CITRIC ACID, Monofunctional chorismate mutase | | Authors: | Khatanbaatar, T, Cordara, G, Krengel, U. | | Deposit date: | 2023-06-30 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural analysis of chorismate mutase and cyclohexadienyl dehydratase from Pseudomonas aeruginosa
To Be Published
|
|
4FS6
 
 | | Crystal structure of the Z-DNA hexamer CGCGCG at 500 mM CaCl2 | | Descriptor: | CALCIUM ION, CHLORIDE ION, DNA (5'-D(*CP*GP*CP*GP*CP*G)-3') | | Authors: | Chatake, T, Sunami, T. | | Deposit date: | 2012-06-27 | | Release date: | 2013-05-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Direct interactions between Z-DNA and alkaline earth cations, discovered in the presence of high concentrations of MgCl2 and CaCl2
J.Inorg.Biochem., 124, 2013
|
|
6YGT
 
 | |
4FS5
 
 | | Crystal structure of the Z-DNA hexamer CGCGCG at 500 mM MgCl2 | | Descriptor: | CHLORIDE ION, DNA (5'-D(*CP*GP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Chatake, T, Sunami, T. | | Deposit date: | 2012-06-26 | | Release date: | 2013-05-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Direct interactions between Z-DNA and alkaline earth cations, discovered in the presence of high concentrations of MgCl2 and CaCl2
J.Inorg.Biochem., 124, 2013
|
|
1EDR
 
 | | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGMO6AATTCGCG) AT 1.6 ANGSTROM | | Descriptor: | 5'-D(*CP*GP*CP*GP*(A47)AP*AP*TP*TP*CP*GP*CP*G)-3', MAGNESIUM ION, SPERMINE | | Authors: | Chatake, T, Hikima, T, Ono, A, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 2000-01-28 | | Release date: | 2000-02-16 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic studies on damaged DNAs. II. N(6)-methoxyadenine can present two alternate faces for Watson-Crick base-pairing, leading to pyrimidine transition mutagenesis.
J.Mol.Biol., 294, 1999
|
|
2Z48
 
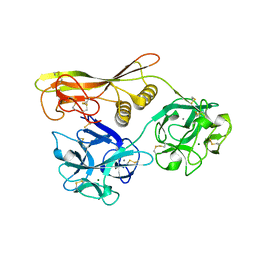 | | Crystal Structure of Hemolytic Lectin CEL-III Complexed with GalNac | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, 2-acetamido-2-deoxy-beta-D-galactopyranose, CALCIUM ION, ... | | Authors: | Hatakeyama, T, Unno, H, Eto, S, Hidemura, H, Uchida, T, Kouzuma, Y. | | Deposit date: | 2007-06-13 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | C-type lectin-like carbohydrate-recognition of the hemolytic lectin CEL-III containing ricin-type beta-trefoil folds
J.Biol.Chem., 282, 2007
|
|
2Z49
 
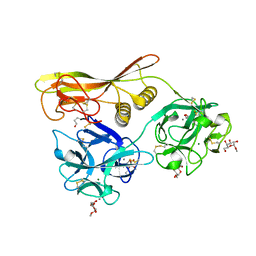 | | Crystal Structure of Hemolytic Lectin CEL-III Complexed with methyl-alpha-D-galactopylanoside | | Descriptor: | CALCIUM ION, Hemolytic lectin CEL-III, MAGNESIUM ION, ... | | Authors: | Hatakeyama, T, Unno, H, Eto, S, Hidemura, H, Uchida, T, Kouzuma, Y. | | Deposit date: | 2007-06-13 | | Release date: | 2007-10-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | C-type lectin-like carbohydrate-recognition of the hemolytic lectin CEL-III containing ricin-type beta-trefoil folds
J.Biol.Chem., 282, 2007
|
|
457D
 
 | | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGMO6AATTCGCG): N6-METHOXYADENOSINE/ THYMIDINE BASE-PAIRS IN B-DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(A47)P*AP*TP*TP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Chatake, T, Ono, A, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 1999-03-06 | | Release date: | 2000-01-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystallographic studies on damaged DNAs. II. N(6)-methoxyadenine can present two alternate faces for Watson-Crick base-pairing, leading to pyrimidine transition mutagenesis.
J.Mol.Biol., 294, 1999
|
|
5D97
 
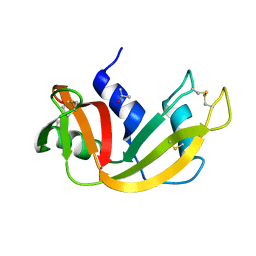 | | Neutron crystal structure of H2O-solvent ribonuclease A | | Descriptor: | ISOPROPYL ALCOHOL, Ribonuclease pancreatic | | Authors: | Chatake, T, Fujiwara, S. | | Deposit date: | 2015-08-18 | | Release date: | 2016-04-06 | | Last modified: | 2018-12-05 | | Method: | NEUTRON DIFFRACTION (1.8 Å) | | Cite: | A technique for determining the deuterium/hydrogen contrast map in neutron macromolecular crystallography
Acta Crystallogr D Struct Biol, 72, 2016
|
|
456D
 
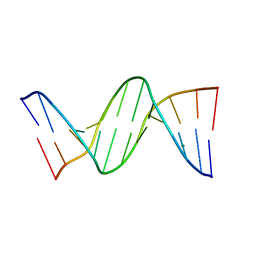 | | MOLECULAR AND CRYSTAL STRUCTURE OF D(CGCGMO6AATCCGCG): THE WATSON-CRICK TYPE N6-METHOXYADENOSINE/CYTIDINE BASE-PAIRS IN B-DNA | | Descriptor: | DNA (5'-D(*CP*GP*CP*GP*(A47)P*AP*TP*CP*CP*GP*CP*G)-3'), MAGNESIUM ION | | Authors: | Chatake, T, Ono, A, Ueno, Y, Matsuda, A, Takenaka, A. | | Deposit date: | 1999-03-06 | | Release date: | 2000-01-01 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystallographic studies on damaged DNAs. I. An N(6)-methoxyadenine residue forms a Watson-Crick pair with a cytosine residue in a B-DNA duplex.
J.Mol.Biol., 294, 1999
|
|
5D6U
 
 | |
7CCO
 
 | | The binding structure of a lanthanide binding tag (LBT3) with lanthanum ion (La3+) | | Descriptor: | LANTHANUM (III) ION, LBT3 | | Authors: | Hatanaka, T, Kikkawa, N, Matsugami, A, Hosokawa, Y, Hayashi, F, Ishida, N. | | Deposit date: | 2020-06-17 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The origins of binding specificity of a lanthanide ion binding peptide.
Sci Rep, 10, 2020
|
|
7CCN
 
 | | The binding structure of a lanthanide binding tag (LBT3) with lutetium ion (Lu3+) | | Descriptor: | LBT3, LUTETIUM (III) ION | | Authors: | Hatanaka, T, Kikkawa, N, Matsugami, A, Hosokawa, Y, Hayashi, F, Ishida, N. | | Deposit date: | 2020-06-17 | | Release date: | 2021-04-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The origins of binding specificity of a lanthanide ion binding peptide.
Sci Rep, 10, 2020
|
|
7FCW
 
 | | X-ray structure of H2O-solvent lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, NICKEL (II) ION | | Authors: | Chatake, T, Tanaka, I, Kusaka, K, Fujiwara, S. | | Deposit date: | 2021-07-15 | | Release date: | 2022-04-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Protonation states of hen egg-white lysozyme observed using D/H contrast neutron crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7FCU
 
 | | X-ray structure of D2O-solvent lysozyme | | Descriptor: | CHLORIDE ION, Lysozyme C, NICKEL (II) ION | | Authors: | Chatake, T, Tanaka, I, Kusaka, K, Fujiwara, S. | | Deposit date: | 2021-07-15 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Protonation states of hen egg-white lysozyme observed using D/H contrast neutron crystallography.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
