5D5Y
 
 | | Structure of Chaetomium thermophilum Skn7 coiled-coil domain, crystal form I | | Descriptor: | Putative transcription factor | | Authors: | Neudegger, T, Verghese, J, Hayer-Hartl, M, Hartl, F.U, Bracher, A. | | Deposit date: | 2015-08-11 | | Release date: | 2015-12-30 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Structure of human heat-shock transcription factor 1 in complex with DNA.
Nat.Struct.Mol.Biol., 23, 2016
|
|
7ZET
 
 | | Crystal structure of human Clusterin, crystal form I | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Clusterin, ... | | Authors: | Yuste-Checa, P, Bracher, A, Hartl, F.U. | | Deposit date: | 2022-03-31 | | Release date: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of human Clusterin, crystal form I
To be published
|
|
7ZEU
 
 | |
1ELR
 
 | | Crystal structure of the TPR2A domain of HOP in complex with the HSP90 peptide MEEVD | | Descriptor: | HSP90-PEPTIDE MEEVD, NICKEL (II) ION, TPR2A-DOMAIN OF HOP | | Authors: | Scheufler, C, Brinker, A, Hartl, F.U, Moarefi, I. | | Deposit date: | 2000-03-14 | | Release date: | 2000-04-26 | | Last modified: | 2017-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine.
Cell(Cambridge,Mass.), 101, 2000
|
|
1ELW
 
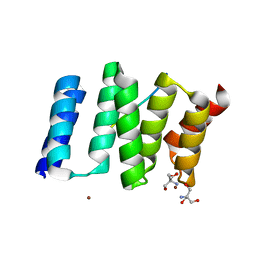 | | Crystal structure of the TPR1 domain of HOP in complex with a HSC70 peptide | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, HSC70-PEPTIDE, NICKEL (II) ION, ... | | Authors: | Scheufler, C, Brinker, A, Hartl, F.U, Moarefi, I. | | Deposit date: | 2000-03-14 | | Release date: | 2000-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of TPR domain-peptide complexes: critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine
Cell(Cambridge,Mass.), 101, 2000
|
|
8QXV
 
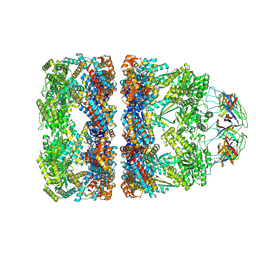 | | In situ structure average of GroEL14-GroES7 complexes with narrow GroEL7 trans ring conformation in Escherichia coli cytosol obtained by cryo electron tomography | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-10-25 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (13.6 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
6HZK
 
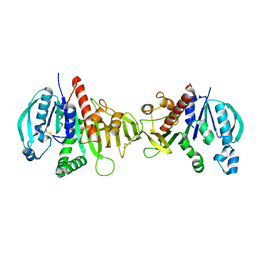 | | Crystal structure of redox-inhibited phosphoribulokinase from Synechococcus sp. (strain PCC 6301) | | Descriptor: | Phosphoribulokinase | | Authors: | Wilson, R.H, Bracher, A, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-10-23 | | Release date: | 2019-03-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of phosphoribulokinase from Synechococcus sp. strain PCC 6301.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6HZL
 
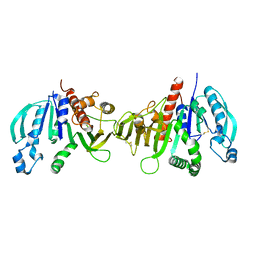 | | Crystal structure of redox-inhibited phosphoribulokinase from Synechococcus sp. (strain PCC 6301), osmate derivative | | Descriptor: | OSMIUM ION, Phosphoribulokinase | | Authors: | Wilson, R.H, Bracher, A, Hartl, F.U, Hayer-Hartl, M. | | Deposit date: | 2018-10-23 | | Release date: | 2019-03-27 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Crystal structure of phosphoribulokinase from Synechococcus sp. strain PCC 6301.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
8QXS
 
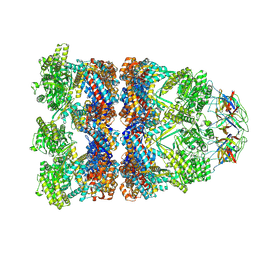 | | CryoEM structure of a GroEL14-GroES7 complex in presence of ADP-BeFx with wide GroEL7 trans ring conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-10-25 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.12 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
8QXU
 
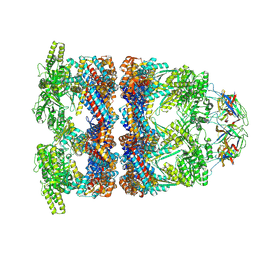 | | In situ structure average of GroEL14-GroES7 complexes with wide GroEL7 trans ring conformation in Escherichia coli cytosol obtained by cryo electron tomography | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-10-25 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (12 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
8QXT
 
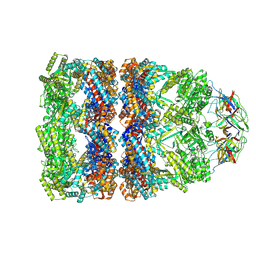 | | CryoEM structure of a GroEL14-GroES7 complex in presence of ADP-BeFx with narrow GroEL7 trans ring conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-10-25 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
1XQS
 
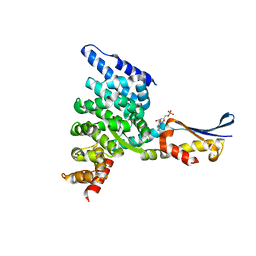 | | Crystal structure of the HspBP1 core domain complexed with the fragment of Hsp70 ATPase domain | | Descriptor: | ADENOSINE MONOPHOSPHATE, HSPBP1 protein, Heat shock 70 kDa protein 1 | | Authors: | Shomura, Y, Dragovic, Z, Chang, H.C, Tzvetkov, N, Young, J.C, Brodsky, J.L, Guerriero, V, Hartl, F.U, Bracher, A. | | Deposit date: | 2004-10-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Regulation of Hsp70 Function by HspBP1; Structural Analysis Reveals an Alternate Mechanism for Hsp70 Nucleotide Exchange
Mol.Cell, 17, 2005
|
|
1XQR
 
 | | Crystal structure of the HspBP1 core domain | | Descriptor: | HspBP1 protein | | Authors: | Shomura, Y, Dragovic, Z, Chang, H.C, Tzvetkov, N, Young, J.C, Brodsky, J.L, Guerriero, V, Hartl, F.U, Bracher, A. | | Deposit date: | 2004-10-13 | | Release date: | 2005-03-01 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Regulation of Hsp70 Function by HspBP1; Structural Analysis Reveals an Alternate Mechanism for Hsp70 Nucleotide Exchange
Mol.Cell, 17, 2005
|
|
6EPC
 
 | | Ground state 26S proteasome (GS2) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (12.3 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
6EPD
 
 | | Substrate processing state 26S proteasome (SPS1) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (15.4 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
6EPF
 
 | | Ground state 26S proteasome (GS1) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11.8 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
6EPE
 
 | | Substrate processing state 26S proteasome (SPS2) | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, 26S proteasome non-ATPase regulatory subunit 11, 26S proteasome non-ATPase regulatory subunit 13, ... | | Authors: | Guo, Q, Lehmer, C, Martinez-Sanchez, A, Rudack, T, Beck, F, Hartmann, H, Hipp, M.S, Hartl, F.U, Edbauer, D, Baumeister, W, Fernandez-Busnadiego, R. | | Deposit date: | 2017-10-11 | | Release date: | 2018-02-07 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (12.8 Å) | | Cite: | In Situ Structure of Neuronal C9orf72 Poly-GA Aggregates Reveals Proteasome Recruitment.
Cell, 172, 2018
|
|
8P4R
 
 | | In situ structure average of GroEL14-GroES14 complexes in Escherichia coli cytosol obtained by cryo electron tomography | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, Co-chaperonin GroES, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (11.9 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
8P4M
 
 | | CryoEM structure of a C7-symmetrical GroEL7-GroES7 cage in presence of ADP-BeFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Beck, F, Bracher, A, Caravajal, A.I, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
8P4O
 
 | | CryoEM structure of a GroEL7-GroES7 cage with encapsulated ordered substrate MetK in the presence of ADP-BeFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Beck, F, Bracher, A, Caravajal, A.I, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (3.04 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
8P4N
 
 | | CryoEM structure of a GroEL7-GroES7 cage with encapsulated disordered substrate MetK in the presence of ADP-BeFx | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, BERYLLIUM TRIFLUORIDE ION, Chaperonin GroEL, ... | | Authors: | Wagner, J, Beck, F, Bracher, A, Caravajal, A.I, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
8P4P
 
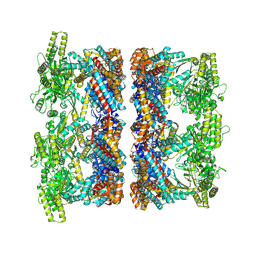 | | Structure average of GroEL14 complexes found in the cytosol of Escherichia coli overexpressing GroEL obtained by cryo electron tomography | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, Chaperonin GroEL, ... | | Authors: | Wagner, J, Caravajal, A.I, Beck, F, Bracher, A, Wan, W, Bohn, S, Koerner, R, Baumeister, W, Fernandez-Busnadiego, R, Hartl, F.U. | | Deposit date: | 2023-05-23 | | Release date: | 2024-07-03 | | Last modified: | 2024-09-25 | | Method: | ELECTRON MICROSCOPY (9.6 Å) | | Cite: | Visualizing chaperonin function in situ by cryo-electron tomography.
Nature, 633, 2024
|
|
6HB3
 
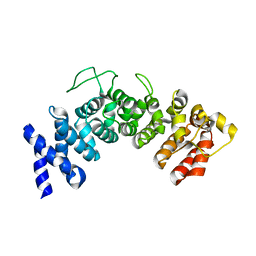 | | Structure of Hgh1, crystal form II | | Descriptor: | Protein HGH1 | | Authors: | Moenkemeyer, L, Klaips, C.L, Balchin, D, Koerner, R, Hartl, F.U, Bracher, A. | | Deposit date: | 2018-08-09 | | Release date: | 2019-02-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Chaperone Function of Hgh1 in the Biogenesis of Eukaryotic Elongation Factor 2.
Mol.Cell, 74, 2019
|
|
6HB2
 
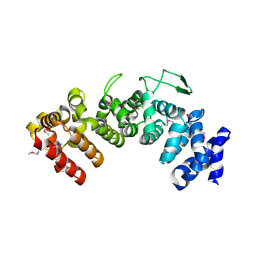 | | Structure of Hgh1, crystal form I, Selenomethionine derivative | | Descriptor: | CHLORIDE ION, Protein HGH1 | | Authors: | Moenkemeyer, L, Klaips, C.L, Balchin, D, Koerner, R, Hartl, F.U, Bracher, A. | | Deposit date: | 2018-08-09 | | Release date: | 2019-02-27 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Chaperone Function of Hgh1 in the Biogenesis of Eukaryotic Elongation Factor 2.
Mol.Cell, 74, 2019
|
|
6HB1
 
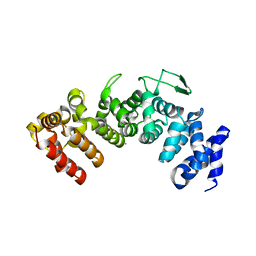 | | Structure of Hgh1, crystal form I | | Descriptor: | CHLORIDE ION, Protein HGH1 | | Authors: | Moenkemeyer, L, Klaips, C.L, Balchin, D, Koerner, R, Hartl, F.U, Bracher, A. | | Deposit date: | 2018-08-09 | | Release date: | 2019-02-27 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Chaperone Function of Hgh1 in the Biogenesis of Eukaryotic Elongation Factor 2.
Mol.Cell, 74, 2019
|
|
