8DCP
 
 | | PI 3-kinase alpha with nanobody 3-126 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD8
 
 | | PI 3-kinase alpha with nanobody 3-142, crosslinked with DSG | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DD4
 
 | | PI 3-kinase alpha with nanobody 3-142 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8DCX
 
 | | PI 3-kinase alpha with nanobody 3-159 | | Descriptor: | Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Hart, J.R, Liu, X, Pan, C, Liang, A, Ueno, L, Xu, Y, Quezada, A, Zou, X, Yang, S, Zhou, Q, Schoonooghe, S, Hassanzadeh-Ghassabeh, G, Xia, T, Shui, W, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-06-17 | | Release date: | 2022-09-21 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Nanobodies and chemical cross-links advance the structural and functional analysis of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6QGW
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with nanobody E6 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NanoE6, Outer membrane protein assembly factor BamA | | Authors: | Hartmann, J.-B, Kaur, H, Jakob, R.P, Zahn, M, Zimmermann, I, Seeger, M, Maier, T, Hiller, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.938 Å) | | Cite: | Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
J.Biomol.Nmr, 73, 2019
|
|
6QGY
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with nanobody B12 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NanoB12, Outer membrane protein assembly factor BamA | | Authors: | Hartmann, J.-B, Kaur, H, Jakob, R.P, Zahn, M, Zimmermann, I, Seeger, M, Maier, T, Hiller, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.509 Å) | | Cite: | Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
J.Biomol.Nmr, 73, 2019
|
|
3SZ7
 
 | |
6QGX
 
 | | Crystal structure of E.coli BamA beta-barrel in complex with nanobody F7 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NanoF7, Outer membrane protein assembly factor BamA | | Authors: | Hartmann, J.-B, Kaur, H, Jakob, R.P, Zahn, M, Zimmermann, I, Seeger, M, Maier, T, Hiller, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
J.Biomol.Nmr, 73, 2019
|
|
8RI4
 
 | | Crystal structure of the SARS-CoV-2 Main Protease inhibited by (2-methylsulfanyl-6,7-dihydro-[1,4]dioxino[2,3-f]benzimidazol-3-yl)-(p-tolyl)methanone | | Descriptor: | 3C-like proteinase nsp5, 4-METHYLBENZOIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Charton, J, Deprez, B, Hanoulle, X. | | Deposit date: | 2023-12-18 | | Release date: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the SARS-CoV-2 Main Protease inhibited by (2-methylsulfanyl-6,7-dihydro-[1,4]dioxino[2,3-f]benzimidazol-3-yl)-(p-tolyl)methanone
To Be Published
|
|
3PSG
 
 | |
3LKU
 
 | |
3VEJ
 
 | | Crystal structure of the Get5 carboxyl domain from S. cerevisiae | | Descriptor: | PHOSPHATE ION, Ubiquitin-like protein MDY2 | | Authors: | Chartron, J.W, Vandervelde, D.G, Rao, M, Clemons Jr, W.M. | | Deposit date: | 2012-01-08 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Get5 Carboxyl-terminal Domain Is a Novel Dimerization Motif That Tethers an Extended Get4/Get5 Complex.
J.Biol.Chem., 287, 2012
|
|
4GOF
 
 | |
4GOE
 
 | |
4GOD
 
 | | Crystal structure of the SGTA homodimerization domain | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Small glutamine-rich tetratricopeptide repeat-containing protein alpha | | Authors: | Chartron, J.W, VanderVelde, D.G, Clemons Jr, W.M. | | Deposit date: | 2012-08-19 | | Release date: | 2012-11-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structures of the Sgt2/SGTA Dimerization Domain with the Get5/UBL4A UBL Domain Reveal an Interaction that Forms a Conserved Dynamic Interface.
Cell Rep, 2, 2012
|
|
4GOC
 
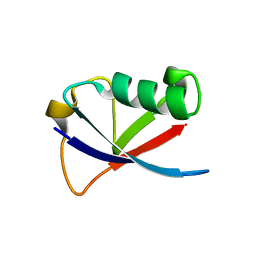 | |
2OG2
 
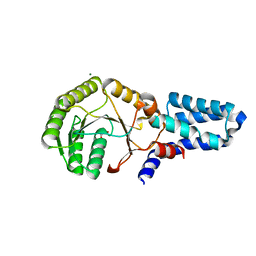 | | Crystal structure of chloroplast FtsY from Arabidopsis thaliana | | Descriptor: | MAGNESIUM ION, MALONATE ION, Putative signal recognition particle receptor | | Authors: | Chartron, J, Chandrasekar, S, Ampornpan, P.J, Shan, S. | | Deposit date: | 2007-01-04 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the Chloroplast Signal Recognition Particle (SRP) Receptor: Domain Arrangement Modulates SRP-Receptor Interaction.
J.Mol.Biol., 375, 2007
|
|
2LNZ
 
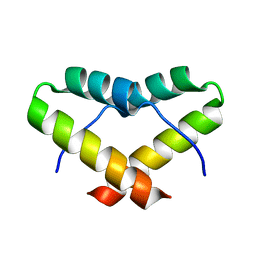 | |
2LXB
 
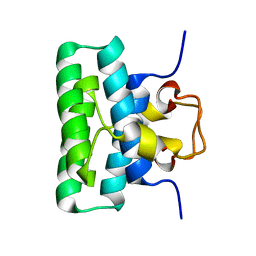 | |
2LXA
 
 | |
2O8V
 
 | | PAPS reductase in a covalent complex with thioredoxin C35A | | Descriptor: | Phosphoadenosine phosphosulfate reductase, Thioredoxin 1 | | Authors: | Chartron, J, Shiau, C, Stout, C.D, Carroll, K.S. | | Deposit date: | 2006-12-12 | | Release date: | 2007-03-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | 3'-Phosphoadenosine-5'-phosphosulfate Reductase in Complex with Thioredoxin: A Structural Snapshot in the Catalytic Cycle.
Biochemistry, 46, 2007
|
|
2GOY
 
 | | Crystal structure of assimilatory adenosine 5'-phosphosulfate reductase with bound APS | | Descriptor: | ADENOSINE-5'-PHOSPHOSULFATE, IRON/SULFUR CLUSTER, adenosine phosphosulfate reductase | | Authors: | Chartron, J, Carroll, K.S, Shiau, C, Gao, H, Leary, J.A, Bertozzi, C.R, Stout, C.D. | | Deposit date: | 2006-04-14 | | Release date: | 2006-09-19 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Substrate Recognition, Protein Dynamics, and Iron-Sulfur Cluster in Pseudomonas aeruginosa Adenosine 5'-Phosphosulfate Reductase.
J.Mol.Biol., 364, 2006
|
|
2LXC
 
 | |
2LO0
 
 | |
8GUB
 
 | | Cryo-EM structure of cancer-specific PI3Kalpha mutant H1047R in complex with BYL-719 | | Descriptor: | (2S)-N~1~-{4-methyl-5-[2-(1,1,1-trifluoro-2-methylpropan-2-yl)pyridin-4-yl]-1,3-thiazol-2-yl}pyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Liu, X, Zhou, Q, Hart, J.R, Xu, Y, Yang, S, Yang, D, Vogt, P.K, Wang, M.-W. | | Deposit date: | 2022-09-11 | | Release date: | 2022-11-23 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Cryo-EM structures of cancer-specific helical and kinase domain mutations of PI3K alpha.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
