4FHW
 
 | |
2P5T
 
 | |
1C6S
 
 | | THE SOLUTION STRUCTURE OF CYTOCHROME C6 FROM THE THERMOPHILIC CYANOBACTERIUM SYNECHOCOCCUS ELONGATUS, NMR, 20 STRUCTURES | | Descriptor: | CYTOCHROME C6, HEME C | | Authors: | Roesch, P, Beissinger, M, Sticht, H, Sutter, M, Ejchart, A, Haehnel, W. | | Deposit date: | 1997-03-31 | | Release date: | 1998-04-08 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome c6 from the thermophilic cyanobacterium Synechococcus elongatus.
EMBO J., 17, 1998
|
|
4FH3
 
 | |
4FHX
 
 | |
2LP2
 
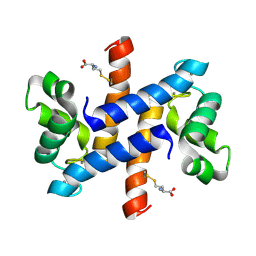 | | Solution structure and dynamics of human S100A1 protein modified at cysteine 85 with homocysteine disulfide bond formation in calcium saturated form | | Descriptor: | 2-AMINO-4-MERCAPTO-BUTYRIC ACID, CALCIUM ION, Protein S100-A1 | | Authors: | Nowakowski, M.E, Jaremko, L, Jaremko, M, Zdanowski, K, Ejchart, A. | | Deposit date: | 2012-01-31 | | Release date: | 2013-02-20 | | Last modified: | 2024-04-03 | | Method: | SOLUTION NMR | | Cite: | Impact of calcium binding and thionylation of S100A1 protein on its nuclear magnetic resonance-derived structure and backbone dynamics.
Biochemistry, 52, 2013
|
|
4FHV
 
 | |
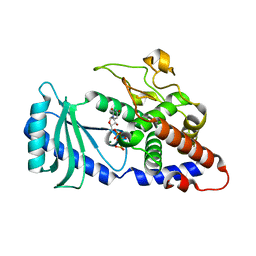
4FH5
 
 | |
4FHY
 
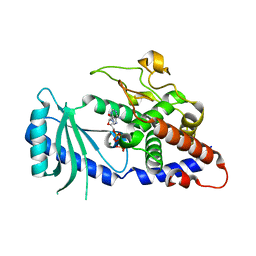 | |
4FGI
 
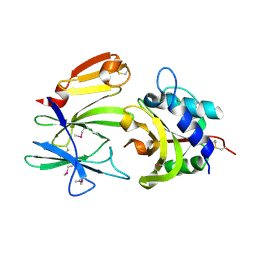 | | Structure of the effector - immunity system Tse1 / Tsi1 from Pseudomonas aeruginosa | | Descriptor: | Tse1, Tsi1 | | Authors: | Benz, J, Sendlmeier, C, Barends, T.R.M, Meinhart, A. | | Deposit date: | 2012-06-04 | | Release date: | 2012-06-20 | | Last modified: | 2012-07-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural Insights into the Effector - Immunity System Tse1/Tsi1 from Pseudomonas aeruginosa.
Plos One, 7, 2012
|
|
2LP3
 
 | | Solution structure of S100A1 Ca2+ | | Descriptor: | CALCIUM ION, Protein S100-A1 | | Authors: | Budzinska, M, Ruszczynska-Bartnik, K, Belczyk-Ciesielska, A, Bierzynski, A, Ejchart, A. | | Deposit date: | 2012-01-31 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Impact of calcium binding and thionylation of S100A1 protein on its nuclear magnetic resonance-derived structure and backbone dynamics.
Biochemistry, 52, 2013
|
|
2M3W
 
 | |
2L0P
 
 | | Solution structure of human apo-S100A1 protein by NMR spectroscopy | | Descriptor: | S100 calcium binding protein A1 | | Authors: | Nowakowski, M, Jaremko, L, Jaremko, M, Bierzynski, A, Zhukov, I, Ejchart, A. | | Deposit date: | 2010-07-12 | | Release date: | 2011-04-20 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure and dynamics of human apo-S100A1 protein.
J.Struct.Biol., 174, 2011
|
|
2LZJ
 
 | | Refined solution structure and dynamics of First Catalytic Cysteine Half-domain from mouse E1 enzyme | | Descriptor: | Ubiquitin-like modifier-activating enzyme 1 | | Authors: | Jaremko, M, Jaremko, L, Nowakowski, M, Szczepanowski, R.H, Filipek, R, Wojciechowski, M, Bochtler, M, Ejchart, A. | | Deposit date: | 2012-10-03 | | Release date: | 2013-09-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | NMR structural studies of the first catalytic half-domain of ubiquitin activating enzyme.
J.Struct.Biol., 185, 2014
|
|
2JPT
 
 | |
2LHL
 
 | | Chemical Shift Assignments and solution structure of human apo-S100A1 E32Q mutant | | Descriptor: | Protein S100-A1 | | Authors: | Ruszczynska-Bartnik, K, Zdanowski, K, Zhukov, I, Bierzynski, A, Ejchart, A. | | Deposit date: | 2011-08-12 | | Release date: | 2012-08-01 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N NMR sequence-specific resonance assignments and relaxation parameters for human apo-S100A1 E32Q mutant
To be Published
|
|
2LLS
 
 | | solution structure of human apo-S100A1 C85M | | Descriptor: | Protein S100-A1 | | Authors: | Budzinska, M, Jaremko, L, Jaremko, M, Zdanowski, K, Zhukov, I, Bierzynski, A, Ejchart, A. | | Deposit date: | 2011-11-17 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Chemical Shift Assignments and solution structure of human apo-S100A1 C85M mutant
To be Published
|
|
2L0I
 
 | | Solution structure of Rtt103 CTD-interacting domain bound to a Ser2 phosphorylated CTD peptide | | Descriptor: | DNA-directed RNA polymerase, Regulator of Ty1 transposition protein 103 | | Authors: | Lunde, B.M, Reichow, S.L, Kim, M, Suh, H, Leeper, T.C, Yang, F, Mutschler, H, Buratowski, S, Meinhart, A, Varani, G. | | Deposit date: | 2010-07-06 | | Release date: | 2010-09-08 | | Last modified: | 2011-07-13 | | Method: | SOLUTION NMR | | Cite: | Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain.
Nat.Struct.Mol.Biol., 17, 2010
|
|
2LUX
 
 | |
2M1I
 
 | |
2KM4
 
 | | Solution structure of Rtt103 CTD interacting domain | | Descriptor: | Regulator of Ty1 transposition protein 103 | | Authors: | Lunde, B.M, Reichow, S, Kim, M, Leeper, T.C, Becker, R, Buratowski, S, Meinhart, A, Varani, G. | | Deposit date: | 2009-07-20 | | Release date: | 2010-09-08 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Cooperative interaction of transcription termination factors with the RNA polymerase II C-terminal domain.
Nat.Struct.Mol.Biol., 17, 2010
|
|
