3KZ4
 
 | | Crystal Structure of the Rotavirus Double Layered Particle | | Descriptor: | Inner capsid protein VP2, Intermediate capsid protein VP6, ZINC ION | | Authors: | Mcclain, B, Settembre, E.C, Bellamy, A.R, Harrison, S.C. | | Deposit date: | 2009-12-07 | | Release date: | 2010-03-02 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | X-ray crystal structure of the rotavirus inner capsid particle at 3.8 A resolution.
J.Mol.Biol., 397, 2010
|
|
6XPQ
 
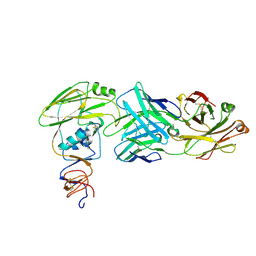 | |
6XPX
 
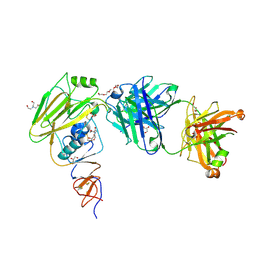 | |
6XPY
 
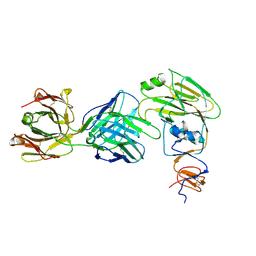 | |
6XQ4
 
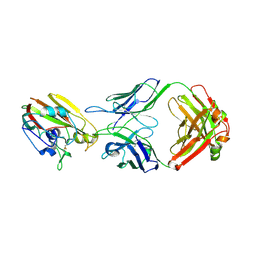 | |
6XPR
 
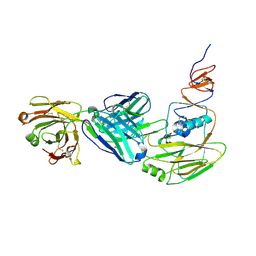 | | Human antibody D2 H1-1/H3-1 H3 in complex with the influenza hemagglutinin head domain of A/Texas/50/2012(H3N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | McCarthy, K.R, Harrison, S.C, Lee, J. | | Deposit date: | 2020-07-08 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (4.092 Å) | | Cite: | A Prevalent Focused Human Antibody Response to the Influenza Virus Hemagglutinin Head Interface.
Mbio, 12, 2021
|
|
6XPO
 
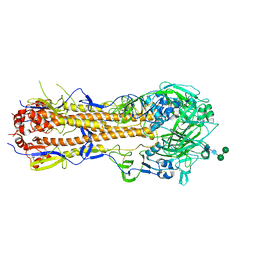 | | Influenza hemagglutinin A/Bangkok/01/1979(H3N2) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin, ... | | Authors: | McCarthy, K.R, Harrison, S.C. | | Deposit date: | 2020-07-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A Prevalent Focused Human Antibody Response to the Influenza Virus Hemagglutinin Head Interface.
Mbio, 12, 2021
|
|
6XPZ
 
 | |
6XQ0
 
 | |
6XQ2
 
 | |
3ML6
 
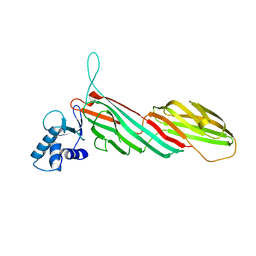 | | a complex between Dishevelled2 and clathrin adaptor AP-2 | | Descriptor: | Chimeric complex between protein Dishevelled2 homolog dvl-2 and clathrin adaptor AP-2 complex subunit mu | | Authors: | Yu, A, Xing, Y, Harrison, S.C, Kirchhausen, T.L. | | Deposit date: | 2010-04-16 | | Release date: | 2010-08-11 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structural analysis of the interaction between Dishevelled2 and clathrin AP-2 adaptor, a critical step in noncanonical Wnt signaling.
Structure, 18, 2010
|
|
3N4S
 
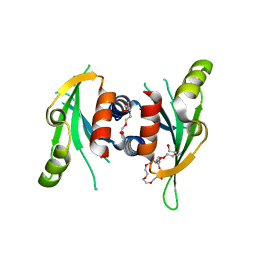 | | Structure of Csm1 C-terminal domain, P21212 form | | Descriptor: | Monopolin complex subunit CSM1, PENTAETHYLENE GLYCOL | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-22 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
3N4R
 
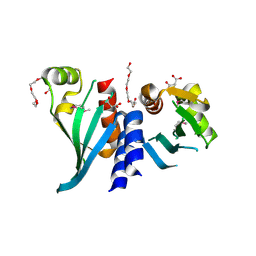 | | Structure of Csm1 C-terminal domain, R3 form | | Descriptor: | MALONATE ION, Monopolin complex subunit CSM1, PENTAETHYLENE GLYCOL | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-22 | | Release date: | 2010-09-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.602 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
3N7N
 
 | | Structure of Csm1/Lrs4 complex | | Descriptor: | Monopolin complex subunit CSM1, Monopolin complex subunit LRS4 | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-27 | | Release date: | 2010-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
3N4X
 
 | | Structure of Csm1 full-length | | Descriptor: | Monopolin complex subunit CSM1 | | Authors: | Corbett, K.D, Harrison, S.C. | | Deposit date: | 2010-05-23 | | Release date: | 2010-09-01 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.408 Å) | | Cite: | The Monopolin Complex Crosslinks Kinetochore Components to Regulate Chromosome-Microtubule Attachments.
Cell(Cambridge,Mass.), 142, 2010
|
|
6XCJ
 
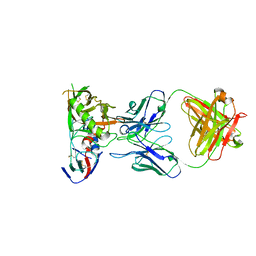 | | Crystal Structure of DH650 Fab from a Rhesus Macaque in Complex with HIV-1 gp120 Core | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, DH650 Fab Heavy Chain, DH650 Fab Light Chain, ... | | Authors: | Raymond, D.D, Chug, H, Harrison, S.C. | | Deposit date: | 2020-06-08 | | Release date: | 2020-12-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Recapitulation of HIV-1 Env-antibody coevolution in macaques leading to neutralization breadth.
Science, 371, 2021
|
|
2YFV
 
 | |
3GZT
 
 | | VP7 recoated rotavirus DLP | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Outer capsid glycoprotein VP7 | | Authors: | Chen, J.Z, Settembre, E.C, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2009-04-07 | | Release date: | 2009-07-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2YFW
 
 | |
3IYJ
 
 | |
3GZU
 
 | | VP7 recoated rotavirus DLP | | Descriptor: | Inner capsid protein VP2, Intermediate capsid protein VP6, ZINC ION | | Authors: | Chen, J.Z, Settembre, E.C, Harrison, S.C, Grigorieff, N. | | Deposit date: | 2009-04-07 | | Release date: | 2009-07-14 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Molecular interactions in rotavirus assembly and uncoating seen by high-resolution cryo-EM.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
6UPH
 
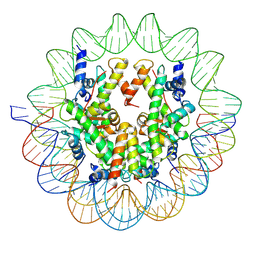 | | Structure of a Yeast Centromeric Nucleosome at 2.7 Angstrom resolution | | Descriptor: | DNA (119-MER), Histone H2A, Histone H2B.1, ... | | Authors: | Migl, D, Kschonsak, M, Arthur, C.P, Khin, Y, Harrison, S.C, Ciferri, C, Dimitrova, Y.N. | | Deposit date: | 2019-10-17 | | Release date: | 2019-11-06 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Cryoelectron Microscopy Structure of a Yeast Centromeric Nucleosome at 2.7 angstrom Resolution.
Structure, 28, 2020
|
|
6V05
 
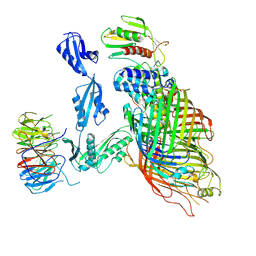 | | Cryo-EM structure of a substrate-engaged Bam complex | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA,Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Tomasek, D, Rawson, S, Lee, J, Wzorek, J.S, Harrison, S.C, Li, Z, Kahne, D. | | Deposit date: | 2019-11-18 | | Release date: | 2020-06-10 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structure of a nascent membrane protein as it folds on the BAM complex.
Nature, 583, 2020
|
|
6WXF
 
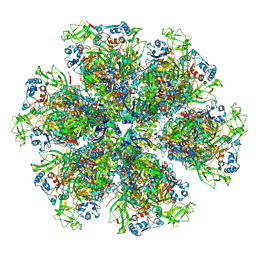 | | Cryo-EM reconstruction of VP5*/VP8* assembly from rhesus rotavirus particles - Intermediate conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Intermediate capsid protein VP6, ... | | Authors: | Herrmann, T, Harrison, S.C, Jenni, S. | | Deposit date: | 2020-05-10 | | Release date: | 2021-01-20 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Functional refolding of the penetration protein on a non-enveloped virus.
Nature, 590, 2021
|
|
6WXG
 
 | | Cryo-EM reconstruction of VP5*/VP8* assembly from rhesus rotavirus particles - Reversed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Intermediate capsid protein VP6, ... | | Authors: | Herrmann, T, Harrison, S.C, Jenni, S. | | Deposit date: | 2020-05-10 | | Release date: | 2021-01-20 | | Last modified: | 2021-03-10 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Functional refolding of the penetration protein on a non-enveloped virus.
Nature, 590, 2021
|
|
