2TBV
 
 | |
3N0A
 
 | | Crystal structure of auxilin (40-400) | | Descriptor: | CALCIUM ION, CHLORIDE ION, Tyrosine-protein phosphatase auxilin | | Authors: | Harrison, S.C, Guan, R, Dai, H, Kirchhausen, T. | | Deposit date: | 2010-05-13 | | Release date: | 2010-09-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the PTEN-like Region of Auxilin, a Detector of Clathrin-Coated Vesicle Budding.
Structure, 18, 2010
|
|
1R69
 
 | | STRUCTURE OF THE AMINO-TERMINAL DOMAIN OF PHAGE 434 REPRESSOR AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | REPRESSOR PROTEIN CI | | Authors: | Mondragon, A, Subbiah, S, Alamo, S.C, Drottar, M, Harrison, S.C. | | Deposit date: | 1988-12-08 | | Release date: | 1989-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the amino-terminal domain of phage 434 repressor at 2.0 A resolution.
J.Mol.Biol., 205, 1989
|
|
5W94
 
 | |
3KAS
 
 | | Machupo virus GP1 bound to human transferrin receptor 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Glycoprotein, ... | | Authors: | Abraham, J, Corbett, K.D, Harrison, S.C. | | Deposit date: | 2009-10-19 | | Release date: | 2010-03-09 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for receptor recognition by New World hemorrhagic fever arenaviruses.
Nat.Struct.Mol.Biol., 17, 2010
|
|
8G0P
 
 | |
8G0Q
 
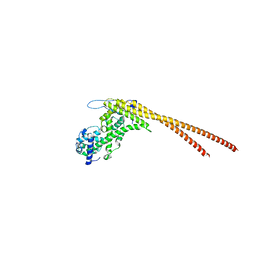 | |
8UDG
 
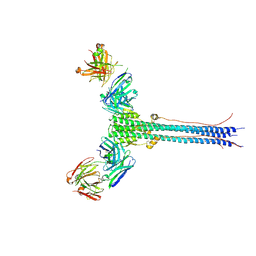 | | S1V2-72 Fab bound to EHA2 from influenza B/Malaysia/2506/2004 | | Descriptor: | Hemagglutinin, S1V2-72 heavy chain, S1V2-72 light chain | | Authors: | Finney, J, Kong, S, Walsh Jr, R.M, Harrison, S.C, Kelsoe, G. | | Deposit date: | 2023-09-28 | | Release date: | 2023-11-15 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.98 Å) | | Cite: | Protective human antibodies against a conserved epitope in pre- and postfusion influenza hemagglutinin.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8V11
 
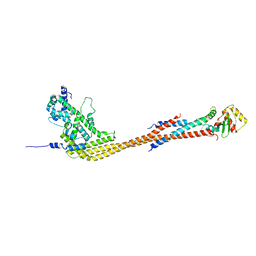 | |
8V10
 
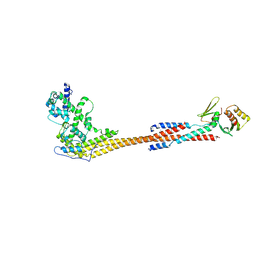 | |
8UK3
 
 | | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2+ (class 6 reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Outer capsid glycoprotein VP7, ... | | Authors: | De Sautu, M, Herrmann, T, Jenni, S, Harrison, S.C. | | Deposit date: | 2023-10-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2.
Plos Pathog., 20, 2024
|
|
8UK2
 
 | | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2+ (class 5 reconstruction) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Outer capsid glycoprotein VP7, ... | | Authors: | De Sautu, M, Herrmann, T, Jenni, S, Harrison, S.C. | | Deposit date: | 2023-10-12 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (8 Å) | | Cite: | The rotavirus VP5*/VP8* conformational transition permeabilizes membranes to Ca2.
Plos Pathog., 20, 2024
|
|
6E4X
 
 | |
6U1X
 
 | | Structure of the Vesicular Stomatitis Virus L Protein in Complex with Its Phosphoprotein Cofactor (3.0 A resolution) | | Descriptor: | Phosphoprotein, RNA-directed RNA polymerase L, ZINC ION | | Authors: | Jenni, S, Bloyet, L.M, Dias-Avalos, R, Liang, B, Wheelman, S.P.J, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2019-08-17 | | Release date: | 2020-01-22 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the Vesicular Stomatitis Virus L Protein in Complex with Its Phosphoprotein Cofactor.
Cell Rep, 30, 2020
|
|
6UEB
 
 | |
6E56
 
 | |
6E8W
 
 | | MPER-TM Domain of HIV-1 envelope glycoprotein (Env) | | Descriptor: | Envelope glycoprotein gp160 | | Authors: | Fu, Q, Shaik, M.M, Cai, Y, Ghantous, F, Piai, A, Peng, H, Rits-Volloch, S, Liu, Z, Harrison, S.C, Seaman, M.S, Chen, B, Chou, J.J. | | Deposit date: | 2018-07-31 | | Release date: | 2018-09-05 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the membrane proximal external region of HIV-1 envelope glycoprotein.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6ULC
 
 | | Structure of full-length, fully glycosylated, non-modified HIV-1 gp160 bound to PG16 Fab at a nominal resolution of 4.6 Angstrom | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Pan, J, Chen, B, Harrison, S.C. | | Deposit date: | 2019-10-07 | | Release date: | 2020-04-29 | | Last modified: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Cryo-EM Structure of Full-length HIV-1 Env Bound With the Fab of Antibody PG16.
J.Mol.Biol., 432, 2020
|
|
3ZXA
 
 | |
3ZXU
 
 | |
7SWO
 
 | | C98C7 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction) | | Descriptor: | C98C7 Fab heavy chain, C98C7 Fab light chain, Spike protein S1 | | Authors: | Windsor, I.W, Tong, P, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-11-20 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Antibodies induced by an ancestral SARS-CoV-2 strain that cross-neutralize variants from Alpha to Omicron BA.1.
Sci Immunol, 7, 2022
|
|
7SWP
 
 | | G32Q4 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction) | | Descriptor: | G32Q4 Fab heavy chain, G32Q4 Fab light chain, Spike protein S1 | | Authors: | Windsor, I.W, Tong, P, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-11-20 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Antibodies induced by an ancestral SARS-CoV-2 strain that cross-neutralize variants from Alpha to Omicron BA.1.
Sci Immunol, 7, 2022
|
|
7SWN
 
 | | G32A4 Fab in complex with SARS-CoV-2 Spike 6P (RBD local reconstruction) | | Descriptor: | G32A4 Fab heavy chain, G32A4 Fab light chain, Spike protein S1 | | Authors: | Windsor, I.W, Tong, P, Wesemann, D.R, Harrison, S.C. | | Deposit date: | 2021-11-20 | | Release date: | 2022-04-27 | | Last modified: | 2022-11-09 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Antibodies induced by an ancestral SARS-CoV-2 strain that cross-neutralize variants from Alpha to Omicron BA.1.
Sci Immunol, 7, 2022
|
|
1HMV
 
 | | THE STRUCTURE OF UNLIGANDED REVERSE TRANSCRIPTASE FROM THE HUMAN IMMUNODEFICIENCY VIRUS TYPE 1 | | Descriptor: | HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P51), HIV-1 REVERSE TRANSCRIPTASE (SUBUNIT P66), MAGNESIUM ION | | Authors: | Rodgers, D.W, Gamblin, S.J, Harris, B.A, Ray, S, Culp, J.S, Hellmig, B, Woolf, D.J, Debouck, C, Harrison, S.C. | | Deposit date: | 1994-12-15 | | Release date: | 1995-03-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1.
Proc.Natl.Acad.Sci.USA, 92, 1995
|
|
8T0P
 
 | | Structure of Cse4 bound to Ame1 and Okp1 | | Descriptor: | Histone H3-like centromeric protein CSE4, Inner kinetochore subunit AME1, Inner kinetochore subunit OKP1, ... | | Authors: | Deng, S, Harrison, S.C. | | Deposit date: | 2023-06-01 | | Release date: | 2023-09-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Recognition of centromere-specific histone Cse4 by the inner kinetochore Okp1-Ame1 complex.
Embo Rep., 24, 2023
|
|
