4Z35
 
 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO-9910539 | | 分子名称: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 3-{1-[(2S,3S)-3-(4-acetyl-3,5-dimethoxyphenyl)-2-(2,3-dihydro-1H-inden-2-ylmethyl)-3-hydroxypropyl]-4-(methoxycarbonyl)-1H-pyrrol-3-yl}propanoic acid, Lysophosphatidic acid receptor 1,Soluble cytochrome b562 | | 著者 | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | 登録日 | 2015-03-30 | | 公開日 | 2015-06-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
4Z36
 
 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO-3080573 | | 分子名称: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, 1-(4-{[(2S,3R)-2-(2,3-dihydro-1H-inden-2-yloxy)-3-(3,5-dimethoxy-4-methylphenyl)-3-hydroxypropyl]oxy}phenyl)cyclopropanecarboxylic acid, Lysophosphatidic acid receptor 1,Soluble cytochrome b562 | | 著者 | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | 登録日 | 2015-03-30 | | 公開日 | 2015-06-03 | | 最終更新日 | 2015-07-01 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
4Z34
 
 | | Crystal Structure of Human Lysophosphatidic Acid Receptor 1 in complex with ONO9780307 | | 分子名称: | (2S)-2,3-dihydroxypropyl (7Z)-tetradec-7-enoate, Lysophosphatidic acid receptor 1, Soluble cytochrome b562, ... | | 著者 | Chrencik, J.E, Roth, C.B, Terakado, M, Kurata, H, Omi, R, Kihara, Y, Warshaviak, D, Nakade, S, Asmar-Rovira, G, Mileni, M, Mizuno, H, Griffith, M.T, Rodgers, C, Han, G.W, Velasquez, J, Chun, J, Stevens, R.C, Hanson, M.A, GPCR Network (GPCR) | | 登録日 | 2015-03-30 | | 公開日 | 2015-06-03 | | 最終更新日 | 2023-09-27 | | 実験手法 | X-RAY DIFFRACTION (3 Å) | | 主引用文献 | Crystal Structure of Antagonist Bound Human Lysophosphatidic Acid Receptor 1.
Cell, 161, 2015
|
|
2DQ6
 
 | | Crystal Structure of Aminopeptidase N from Escherichia coli | | 分子名称: | Aminopeptidase N, SULFATE ION, ZINC ION | | 著者 | Nakajima, Y, Onohara, Y, Ito, K, Yoshimoto, T. | | 登録日 | 2006-05-22 | | 公開日 | 2006-08-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Aminopeptidase N (proteobacteria alanyl aminopeptidase) from Escherichia coli: Crystal structure and conformational change of the methionine 260 residue involved in substrate recognition
J.Biol.Chem., 281, 2006
|
|
1IYX
 
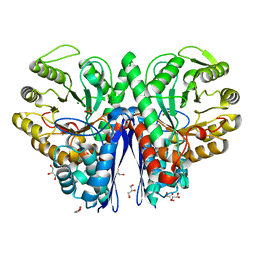 | | Crystal structure of enolase from Enterococcus hirae | | 分子名称: | ENOLASE, GLYCEROL, MAGNESIUM ION, ... | | 著者 | Hosaka, T, Meguro, T, Yamato, I, Shirakihara, Y. | | 登録日 | 2002-09-12 | | 公開日 | 2003-07-29 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.8 Å) | | 主引用文献 | Crystal Structure of Enterococcus hirae Enolase at 2.8 A Resolution
J.BIOCHEM.(TOKYO), 133, 2003
|
|
5CZD
 
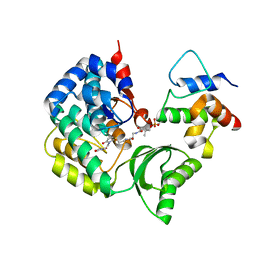 | | The complex structure of VinK with VinL | | 分子名称: | 1,1'-ethane-1,2-diyldipyrrolidine-2,5-dione, 4'-PHOSPHOPANTETHEINE, Acyl-carrier-protein, ... | | 著者 | Miyanaga, A, Iwasawa, S, Shinohara, Y, Kudo, F, Eguchi, T. | | 登録日 | 2015-07-31 | | 公開日 | 2016-02-03 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.34 Å) | | 主引用文献 | Structure-based analysis of the molecular interactions between acyltransferase and acyl carrier protein in vicenistatin biosynthesis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
7DD4
 
 | |
5CZC
 
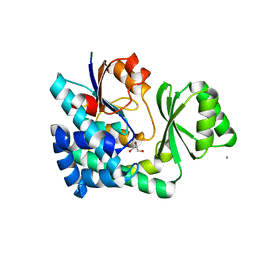 | | The structure of VinK | | 分子名称: | CALCIUM ION, GLYCEROL, Malonyl-CoA-[acyl-carrier-protein] transacylase | | 著者 | Miyanaga, A, Iwasawa, S, Shinohara, Y, Kudo, F, Eguchi, T. | | 登録日 | 2015-07-31 | | 公開日 | 2016-02-03 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Structure-based analysis of the molecular interactions between acyltransferase and acyl carrier protein in vicenistatin biosynthesis.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1V84
 
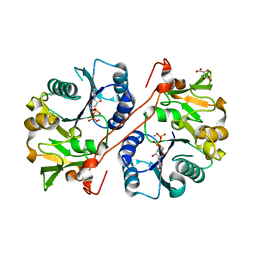 | | Crystal structure of human GlcAT-P in complex with N-acetyllactosamine, Udp, and Mn2+ | | 分子名称: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 1, L(+)-TARTARIC ACID, MANGANESE (II) ION, ... | | 著者 | Kakuda, S, Shiba, T, Ishiguro, M, Tagawa, H, Oka, S, Kajihara, Y, Kawasaki, T, Wakatsuki, S, Kato, R. | | 登録日 | 2003-12-27 | | 公開日 | 2004-05-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.82 Å) | | 主引用文献 | Structural Basis for Acceptor Substrate Recognition of a Human Glucuronyltransferase, GlcAT-P, an Enzyme Critical in the Biosynthesis of the Carbohydrate Epitope HNK-1
J.Biol.Chem., 279, 2004
|
|
1V83
 
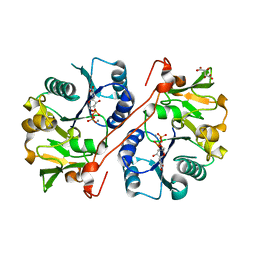 | | Crystal structure of human GlcAT-P in complex with Udp and Mn2+ | | 分子名称: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 1, L(+)-TARTARIC ACID, MANGANESE (II) ION, ... | | 著者 | Kakuda, S, Shiba, T, Ishiguro, M, Tagawa, H, Oka, S, Kajihara, Y, Kawasaki, T, Wakatsuki, S, Kato, R. | | 登録日 | 2003-12-27 | | 公開日 | 2004-05-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structural Basis for Acceptor Substrate Recognition of a Human Glucuronyltransferase, GlcAT-P, an Enzyme Critical in the Biosynthesis of the Carbohydrate Epitope HNK-1
J.Biol.Chem., 279, 2004
|
|
1V82
 
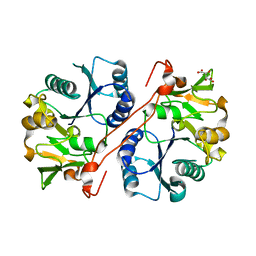 | | Crystal structure of human GlcAT-P apo form | | 分子名称: | Galactosylgalactosylxylosylprotein 3-beta-glucuronosyltransferase 1, L(+)-TARTARIC ACID | | 著者 | Kakuda, S, Shiba, T, Ishiguro, M, Tagawa, H, Oka, S, Kajihara, Y, Kawasaki, T, Wakatsuki, S, Kato, R. | | 登録日 | 2003-12-27 | | 公開日 | 2004-05-25 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.85 Å) | | 主引用文献 | Structural Basis for Acceptor Substrate Recognition of a Human Glucuronyltransferase, GlcAT-P, an Enzyme Critical in the Biosynthesis of the Carbohydrate Epitope HNK-1
J.Biol.Chem., 279, 2004
|
|
2LCF
 
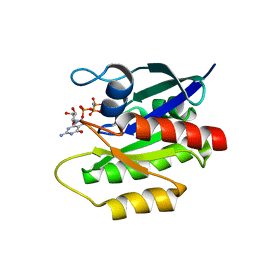 | | Solution structure of GppNHp-bound H-RasT35S mutant protein | | 分子名称: | GTPase HRas, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER | | 著者 | Araki, M, Shima, F, Yoshikawa, Y, Muraoka, S, Ijiri, Y, Nagahara, Y, Shirono, T, Kataoka, T, Tamura, A. | | 登録日 | 2011-04-28 | | 公開日 | 2011-09-28 | | 最終更新日 | 2024-05-01 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Solution structure of the state 1 conformer of GTP-bound H-Ras protein and distinct dynamic properties between the state 1 and state 2 conformers.
J.Biol.Chem., 286, 2011
|
|
3EOQ
 
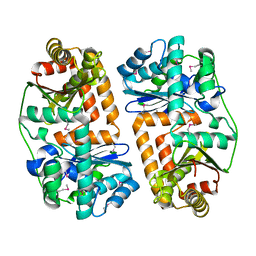 | | The crystal structure of putative zinc protease beta-subunit from Thermus thermophilus HB8 | | 分子名称: | Putative zinc protease | | 著者 | Ohtsuka, J, Ichihara, Y, Ebihara, A, Yokoyama, S, Kuramitsu, S, Nagata, K, Tanokura, M. | | 登録日 | 2008-09-29 | | 公開日 | 2009-03-17 | | 最終更新日 | 2024-10-09 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Crystal structure of TTHA1264, a putative M16-family zinc peptidase from Thermus thermophilus HB8 that is homologous to the beta subunit of mitochondrial processing peptidase.
Proteins, 2009
|
|
2DSK
 
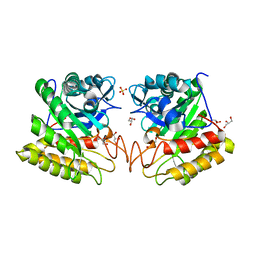 | | Crystal structure of catalytic domain of hyperthermophilic chitinase from Pyrococcus furiosus | | 分子名称: | GLYCEROL, SULFATE ION, chitinase | | 著者 | Nakamura, T, Mine, S, Hagihara, Y, Ishikawa, K, Uegaki, K. | | 登録日 | 2006-06-30 | | 公開日 | 2007-02-06 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structure of the catalytic domain of the hyperthermophilic chitinase from Pyrococcus furiosus
ACTA CRYSTALLOGR.,SECT.F, 63, 2007
|
|
2E2G
 
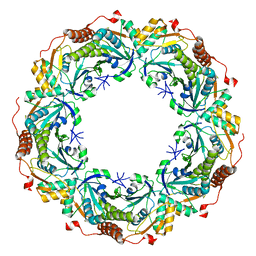 | | Crystal structure of archaeal peroxiredoxin, thioredoxin peroxidase from Aeropyrum pernix K1 (pre-oxidation form) | | 分子名称: | Probable peroxiredoxin | | 著者 | Nakamura, T, Yamamoto, T, Abe, M, Matsumura, H, Hagihara, Y, Goto, T, Yamaguchi, T, Inoue, T. | | 登録日 | 2006-11-13 | | 公開日 | 2007-11-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.4 Å) | | 主引用文献 | Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2CZN
 
 | | Solution structure of the chitin-binding domain of hyperthermophilic chitinase from pyrococcus furiosus | | 分子名称: | chitinase | | 著者 | Uegaki, T, Ikegami, T, Nakamura, T, Hagihara, Y, Mine, S, Inoue, T, Matsumura, H, Ataka, M, Ishikawa, K. | | 登録日 | 2005-07-13 | | 公開日 | 2006-07-18 | | 最終更新日 | 2024-05-29 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Tertiary structure and carbohydrate recognition by the chitin-binding domain of a hyperthermophilic chitinase from Pyrococcus furiosus.
J.Mol.Biol., 381, 2008
|
|
2E2M
 
 | | Crystal structure of archaeal peroxiredoxin, thioredoxin peroxidase from Aeropyrum pernix K1 (sulfinic acid form) | | 分子名称: | Probable peroxiredoxin | | 著者 | Nakamura, T, Yamamoto, T, Abe, M, Matsumura, H, Hagihara, Y, Goto, T, Yamaguchi, T, Inoue, T. | | 登録日 | 2006-11-14 | | 公開日 | 2007-11-20 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
2MST
 
 | | MUSASHI1 RBD2, NMR | | 分子名称: | PROTEIN (MUSASHI1) | | 著者 | Nagata, T, Kanno, R, Kurihara, Y, Uesugi, S, Imai, T, Sakakibara, S, Okano, H, Katahira, M. | | 登録日 | 1999-05-19 | | 公開日 | 2000-05-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure, backbone dynamics and interactions with RNA of the C-terminal RNA-binding domain of a mouse neural RNA-binding protein, Musashi1.
J.Mol.Biol., 287, 1999
|
|
4OIK
 
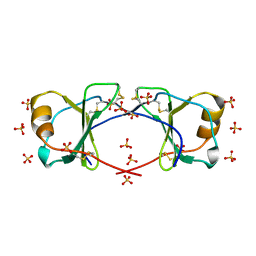 | | (Quasi-)Racemic X-ray crystal structure of glycosylated chemokine Ser-CCL1. | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C-C motif chemokine 1, CITRIC ACID, ... | | 著者 | Okamoto, R, Mandal, K, Sawaya, M.R, Kajihara, Y, Yeates, T.O, Kent, S.B.H. | | 登録日 | 2014-01-19 | | 公開日 | 2014-05-07 | | 最終更新日 | 2023-12-06 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | (Quasi-)Racemic X-ray Structures of Glycosylated and Non-Glycosylated Forms of the Chemokine Ser-CCL1 Prepared by Total Chemical Synthesis.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4OIJ
 
 | | X-ray crystal structure of racemic non-glycosylated chemokine Ser-CCL1 | | 分子名称: | C-C motif chemokine 1, D-Ser-CCL1, SULFATE ION | | 著者 | Okamoto, R, Mandal, K, Sawaya, M.R, Kajihara, Y, Yeates, T.O, Kent, S.B.H. | | 登録日 | 2014-01-19 | | 公開日 | 2014-05-07 | | 最終更新日 | 2014-05-21 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | (Quasi-)Racemic X-ray Structures of Glycosylated and Non-Glycosylated Forms of the Chemokine Ser-CCL1 Prepared by Total Chemical Synthesis.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
2MSS
 
 | | MUSASHI1 RBD2, NMR | | 分子名称: | PROTEIN (MUSASHI1) | | 著者 | Nagata, T, Kanno, R, Kurihara, Y, Uesugi, S, Imai, T, Sakakibara, S, Okano, H, Katahira, M. | | 登録日 | 1999-05-19 | | 公開日 | 2000-05-19 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | Structure, backbone dynamics and interactions with RNA of the C-terminal RNA-binding domain of a mouse neural RNA-binding protein, Musashi1.
J.Mol.Biol., 287, 1999
|
|
3IF5
 
 | | Crystal Structure Analysis of Mglu | | 分子名称: | Salt-tolerant glutaminase | | 著者 | Yoshimune, K, Shirakihara, Y. | | 登録日 | 2009-07-24 | | 公開日 | 2009-08-04 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (2.44 Å) | | 主引用文献 | Crystal structure of salt-tolerant glutaminase from Micrococcus luteus K-3 in the presence and absence of its product L-glutamate and its activator Tris.
Febs J., 277, 2010
|
|
2NVL
 
 | | Crystal structure of archaeal peroxiredoxin, thioredoxin peroxidase from Aeropyrum pernix K1 (sulfonic acid form) | | 分子名称: | Probable peroxiredoxin | | 著者 | Nakamura, T, Yamamoto, T, Abe, M, Matsumura, H, Hagihara, Y, Goto, T, Yamaguchi, T, Inoue, T. | | 登録日 | 2006-11-13 | | 公開日 | 2007-11-20 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.36 Å) | | 主引用文献 | Oxidation of archaeal peroxiredoxin involves a hypervalent sulfur intermediate
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3IHA
 
 | |
3IH8
 
 | |
