2ED4
 
 | | Crystal structure of flavin reductase HpaC complexed with FAD and NAD | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, flavin reductase (HpaC) of 4-hydroxyphenylacetate 3-monooxygenae | | Authors: | Kim, S.H, Hisano, T, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-02-14 | | Release date: | 2008-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8: Structural basis for the flavin affinity
Proteins, 70, 2008
|
|
2IDE
 
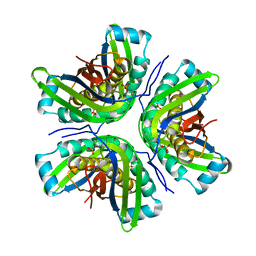 | | Crystal Structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8 | | Descriptor: | Molybdenum cofactor biosynthesis protein C, PHOSPHATE ION | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-09-15 | | Release date: | 2007-09-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the molybdenum cofactor biosynthesis protein C (TTHA1789) from Thermus Theromophilus HB8
To be Published
|
|
2CTZ
 
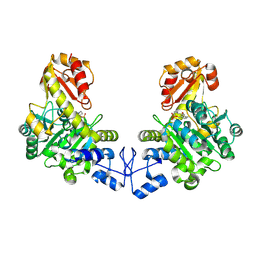 | | Crystal structure of o-acetyl homoserine sulfhydrylase from Thermus thermophilus HB8 | | Descriptor: | O-acetyl-L-homoserine sulfhydrylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Imagawa, T, Kousumi, Y, Tsuge, H, Utsunomiya, H, Ebihara, A, Nakagawa, N, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-05-24 | | Release date: | 2005-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of o-acetyl homoserine sulfhydrylase from Thermus thermophilus HB8
To be Published
|
|
2EIS
 
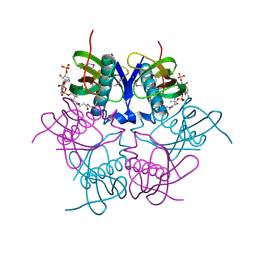 | | X-ray structure of acyl-CoA hydrolase-like protein, TT1379, from Thermus thermophilus HB8 | | Descriptor: | COENZYME A, Hypothetical protein TTHB207 | | Authors: | Kamitori, S, Yoshida, H, Satoh, S, Iino, H, Ebihara, A, Chen, L, Fu, Z.-Q, Chrzas, J, Wang, B.-C, Yokoyama, S, Kuramitsu, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-13 | | Release date: | 2008-03-18 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | X-ray structure of acyl-CoA hydrolase-like protein, TT1379, from Thermus thermophilus HB8
To be Published
|
|
2EHD
 
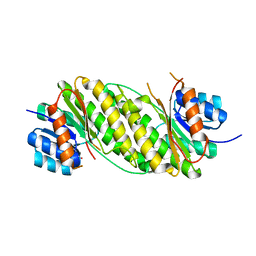 | | Crystal Structure Analysis of Oxidoreductase | | Descriptor: | COBALT (II) ION, Oxidoreductase, short-chain dehydrogenase/reductase family | | Authors: | Kamiya, N, Hikima, T, Ebihara, A, Inoue, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-06 | | Release date: | 2008-03-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure analysis of putative oxidoreductase from Thermus thermophilus HB8
to be published
|
|
2ECU
 
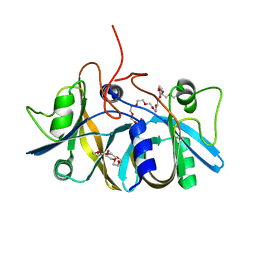 | | Crystal structure of flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, DODECAETHYLENE GLYCOL, flavin reductase (HpaC) of 4-hydroxyphenylacetate 3-monooxygnease | | Authors: | Kim, S.H, Hisano, T, Iwasaki, W, Ebihara, A, Miki, K. | | Deposit date: | 2007-02-14 | | Release date: | 2008-01-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the flavin reductase component (HpaC) of 4-hydroxyphenylacetate 3-monooxygenase from Thermus thermophilus HB8: Structural basis for the flavin affinity
Proteins, 70, 2008
|
|
2E6K
 
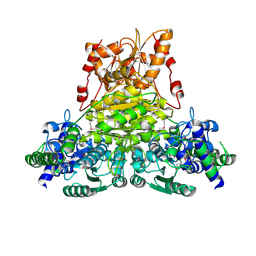 | | X-ray structure of Thermus thermopilus HB8 TT0505 | | Descriptor: | Transketolase | | Authors: | Yoshida, H, Kamitori, S, Agari, Y, Iino, H, Kanagawa, M, Nakagawa, N, Ebihara, A, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-12-27 | | Release date: | 2007-11-13 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | X-ray structure of Thermus thermophilus HB8 TT0505
To be Published
|
|
2PT5
 
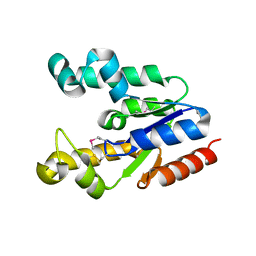 | | Crystal Structure Of Shikimate Kinase (aq_2177) From Aquifex Aeolicus vf5 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Shikimate kinase | | Authors: | Jeyakanthan, J, Nithya, N, Shimada, A, Velmurugan, D, Ebihara, A, Shinkai, A, Kuramitsu, S, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-05-08 | | Release date: | 2008-05-13 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure Of Shikimate Kinase (aq_2177) From Aquifex Aeolicus vf5
To be Published
|
|
2CWW
 
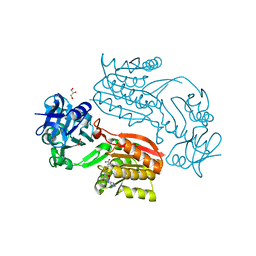 | | Crystal structure of Thermus thermophilus TTHA1280, a putative SAM-dependent RNA methyltransferase, in complex with S-adenosyl-L-homocysteine | | Descriptor: | ACETIC ACID, GLYCEROL, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Pioszak, A.A, Murayama, K, Nakagawa, N, Ebihara, A, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2005-06-27 | | Release date: | 2005-10-11 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of a putative RNA 5-methyluridine methyltransferase, Thermus thermophilus TTHA1280, and its complex with S-adenosyl-L-homocysteine.
Acta Crystallogr.,Sect.F, 61, 2005
|
|
2DQB
 
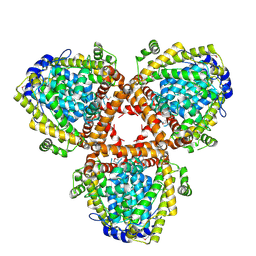 | | Crystal structure of dNTP triphosphohydrolase from Thermus thermophilus HB8, which is homologous to dGTP triphosphohydrolase | | Descriptor: | Deoxyguanosinetriphosphate triphosphohydrolase, putative, MAGNESIUM ION | | Authors: | Kondo, N, Nakagawa, N, Ebihara, A, Chen, L, Liu, Z.-J, Wang, B.-C, Yokoyama, S, Kuramitsu, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2006-05-25 | | Release date: | 2007-01-23 | | Last modified: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of dNTP-inducible dNTP triphosphohydrolase: insight into broad specificity for dNTPs and triphosphohydrolase-type hydrolysis
ACTA CRYSTALLOGR.,SECT.D, 63, 2007
|
|
2EJ5
 
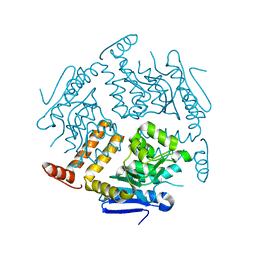 | | Crystal structure of GK2038 protein (enoyl-CoA hydratase subunit II) from Geobacillus kaustophilus | | Descriptor: | Enoyl-CoA hydratase subunit II | | Authors: | Okazaki, N, Agari, Y, Ebihara, A, Chen, L, Fu, Z.Q, Chrzas, J, Wang, B.C, Kuramitsu, S, Yamamoto, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-15 | | Release date: | 2007-09-18 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of GK2038 protein (enoyl-CoA hydratase subunit II) from Geobacillus kaustophilus
To be Published
|
|
4PEO
 
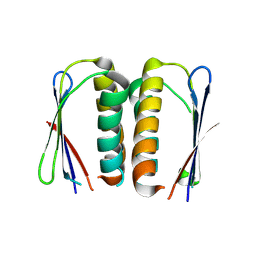 | | Crystal structure of a hypothetical protein from Staphylococcus aureus. | | Descriptor: | Hypothetical protein | | Authors: | McGrath, T.E, Kisselman, G, Romanov, V, Wu-Brown, J, Soloveychik, M, Chan, T.S.Y, Gordon, R.D, Thambipillai, D, Dharamsi, A, Mansoury, K, Battaile, K.P, Edwards, A.M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-04-24 | | Release date: | 2015-05-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Crystal structure of a hypothetical protein from Staphylococcus aureus.
To Be Published
|
|
3EOQ
 
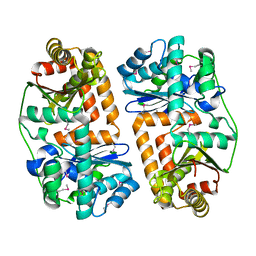 | | The crystal structure of putative zinc protease beta-subunit from Thermus thermophilus HB8 | | Descriptor: | Putative zinc protease | | Authors: | Ohtsuka, J, Ichihara, Y, Ebihara, A, Yokoyama, S, Kuramitsu, S, Nagata, K, Tanokura, M. | | Deposit date: | 2008-09-29 | | Release date: | 2009-03-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Crystal structure of TTHA1264, a putative M16-family zinc peptidase from Thermus thermophilus HB8 that is homologous to the beta subunit of mitochondrial processing peptidase.
Proteins, 2009
|
|
3MCI
 
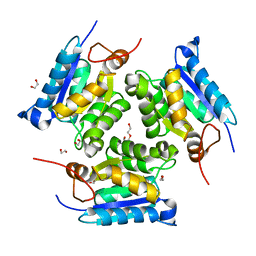 | | Crystal structure of molybdenum cofactor biosynthesis (AQ_061) from aquifex aeolicus VF5 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Molybdenum cofactor biosynthesis MOG | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-03-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures, dynamics and functional implications of molybdenum-cofactor biosynthesis protein MogA from two thermophilic organisms
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3MCH
 
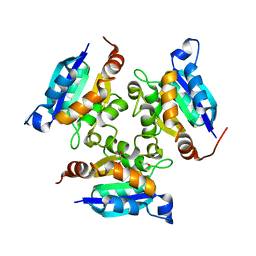 | | Crystal structure of the molybdopterin biosynthesis enzyme MoaB (TTHA0341) from thermus theromophilus HB8 | | Descriptor: | 1,2-ETHANEDIOL, Molybdopterin biosynthesis enzyme, MoaB | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Sekar, K, Baba, S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2010-03-29 | | Release date: | 2011-01-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Crystal structures, dynamics and functional implications of molybdenum-cofactor biosynthesis protein MogA from two thermophilic organisms
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2P5U
 
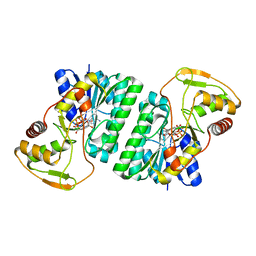 | | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase | | Authors: | Fu, Z.-Q, Chen, L, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhao, M, Dillard, B, Chrzas, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-17 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Crystal structure of Thermus thermophilus HB8 UDP-glucose 4-epimerase complex with NAD
To be Published
|
|
3K8U
 
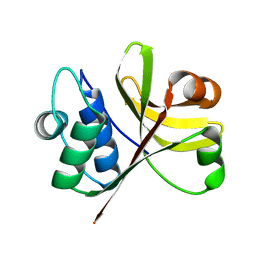 | | Crystal Structure of the Peptidase Domain of Streptococcus ComA, a Bi-functional ABC Transporter Involved in Quorum Sensing Pathway | | Descriptor: | Putative ABC transporter, ATP-binding protein ComA | | Authors: | Ishii, S, Yano, T, Ebihara, A, Okamoto, A, Manzoku, M, Hayashi, H. | | Deposit date: | 2009-10-14 | | Release date: | 2010-02-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the peptidase domain of Streptococcus ComA, a bifunctional ATP-binding cassette transporter involved in the quorum-sensing pathway
J.Biol.Chem., 285, 2010
|
|
2OMD
 
 | | Crystal structure of molybdopterin converting factor subunit 2 (aq_2181) from aquifex aeolicus VF5 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Agari, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-22 | | Release date: | 2008-01-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of molybdopterin converting factor subunit 2 (aq_2181) from aquifex aeolicus VF5
To be Published
|
|
2P8T
 
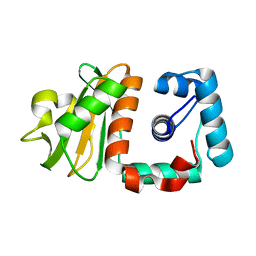 | | Hypothetical protein PH0730 from Pyrococcus horikoshii OT3 | | Descriptor: | Hypothetical protein PH0730 | | Authors: | Chen, L, Zhao, M, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhu, J, Swindell, J.T, Fu, Z.-Q, Chrzas, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-23 | | Release date: | 2007-04-24 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Hypothetical protein PH0730 from Pyrococcus horikoshii OT3
To be Published
|
|
2PCL
 
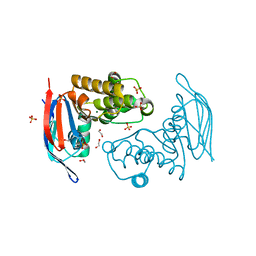 | | Crystal structure of ABC transporter with complex (aq_297) from aquifex aeolicus VF5 | | Descriptor: | 1,2-ETHANEDIOL, Lipoprotein-releasing system ATP-binding protein lolD, MAGNESIUM ION, ... | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Rafi, Z.A, Sekar, K, Ebihara, A, Nakagawa, N, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of ABC transporter with complex (aq_297) from aquifex aeolicus VF5
To be Published
|
|
2PCR
 
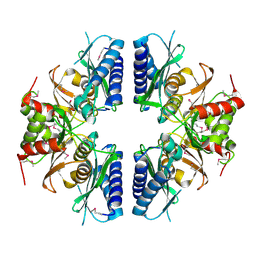 | | Crystal structure of Myo-inositol-1(or 4)-monophosphatase (aq_1983) from Aquifex Aeolicus VF5 | | Descriptor: | Inositol-1-monophosphatase | | Authors: | Jeyakanthan, J, Gayathri, D, Velmurugan, D, Agari, Y, Bessho, Y, Ellis, M.J, Antonyuk, S.V, Strange, R.W, Hasnain, S.S, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Myo-inositol-1(or 4)-monophosphatase (aq_1983) from Aquifex Aeolicus VF5
To be Published
|
|
2P68
 
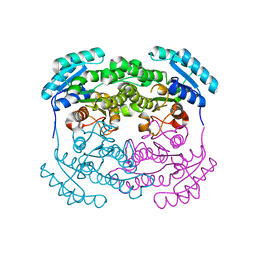 | | Crystal Structure of aq_1716 from Aquifex Aeolicus VF5 | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase | | Authors: | Chen, L, Chen, L.-Q, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Zhao, M, Dillard, B, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure of aq_1716 from Aquifex aeolicus VF5
To be Published
|
|
2P62
 
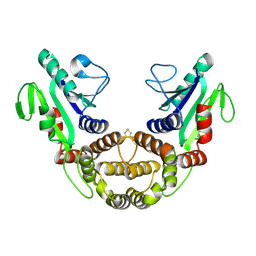 | | Crystal structure of hypothetical protein PH0156 from Pyrococcus horikoshii OT3 | | Descriptor: | Hypothetical protein PH0156 | | Authors: | Fu, Z.-Q, Chen, L, Zhu, J, Swindell, J.T, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Chrzas, J, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-16 | | Release date: | 2007-04-17 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of hypothetical protein PH0156 from Pyrococcus horikoshii OT3
To be Published
|
|
2PCQ
 
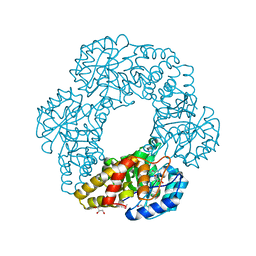 | | Crystal structure of putative dihydrodipicolinate synthase (TTHA0737) from Thermus Thermophilus HB8 | | Descriptor: | GLYCEROL, POTASSIUM ION, Putative dihydrodipicolinate synthase | | Authors: | Jeyakanthan, J, Kanaujia, S.P, Vasuki Ranjani, C, Sekar, K, Kitamura, Y, Ebihara, A, Kuramitsu, S, Shinkai, A, Shiro, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-30 | | Release date: | 2007-10-02 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of putative dihydrodipicolinate synthase (TTHA0737) from Thermus Thermophilus HB8
To be Published
|
|
2P17
 
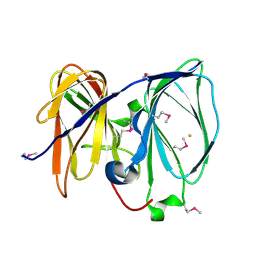 | | Crystal structure of GK1651 from Geobacillus kaustophilus | | Descriptor: | FE (III) ION, Pirin-like protein | | Authors: | Zhu, J, Swindell II, J.T, Chen, L, Ebihara, A, Shinkai, A, Kuramitsu, S, Yokoyama, S, Fu, Z.-Q, Rose, J.P, Wang, B.C, Southeast Collaboratory for Structural Genomics (SECSG), RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-03-02 | | Release date: | 2007-05-01 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal structure of GK1651 from Geobacillus kaustophilus
To be Published
|
|
