4RV0
 
 | | Crystal structure of TN complex | | Descriptor: | Nuclear protein localization protein 4 homolog, SULFATE ION, Transitional endoplasmic reticulum ATPase TER94 | | Authors: | Hao, Q, Jiao, S, Shi, Z.B, Zhou, Z.C. | | Deposit date: | 2014-11-23 | | Release date: | 2015-11-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Crystal structure of TN complex
To be Published
|
|
5F21
 
 | | human CD38 in complex with nanobody MU375 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, GLYCEROL, SULFATE ION, ... | | Authors: | Hao, Q, Zhang, H. | | Deposit date: | 2015-12-01 | | Release date: | 2016-06-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Immuno-targeting the multifunctional CD38 using nanobody
Sci Rep, 6, 2016
|
|
4O27
 
 | | Crystal structure of MST3-MO25 complex with WIF motif | | Descriptor: | 5-mer peptide from serine/threonine-protein kinase 24, ADENOSINE-5'-DIPHOSPHATE, Calcium-binding protein 39, ... | | Authors: | Hao, Q, Feng, M, Zhou, Z.C. | | Deposit date: | 2013-12-16 | | Release date: | 2014-12-03 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.185 Å) | | Cite: | Structural insights into regulatory mechanisms of MO25-mediated kinase activation.
J.Struct.Biol., 186, 2014
|
|
3NS2
 
 | | High-resolution structure of pyrabactin-bound PYL2 | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2 | | Authors: | Hao, Q, Yin, P, Yan, C, Yuan, X, Wang, J, Yan, N. | | Deposit date: | 2010-07-01 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.634 Å) | | Cite: | Single amino acid alteration between Valine and Isoleucine determines the distinct pyrabactin selectivity by PYL1 and PYL2
J.Biol.Chem., 285, 2010
|
|
3RT0
 
 | | Crystal structure of PYL10-HAB1 complex in the absence of abscisic acid (ABA) | | Descriptor: | Abscisic acid receptor PYL10, MAGNESIUM ION, Protein phosphatase 2C 16 | | Authors: | Hao, Q, Yin, P, Li, W, Wang, L, Yan, C, Wang, J, Yan, N. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.113 Å) | | Cite: | The Molecular Basis of ABA-Independent Inhibition of PP2Cs by a Subclass of PYL Proteins
Mol.Cell, 42, 2011
|
|
3RT2
 
 | | Crystal structure of apo-PYL10 | | Descriptor: | Abscisic acid receptor PYL10 | | Authors: | Hao, Q, Yin, P, Li, W, Wang, L, Yan, C, Wang, J, Yan, N. | | Deposit date: | 2011-05-02 | | Release date: | 2011-06-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The Molecular Basis of ABA-Independent Inhibition of PP2Cs by a Subclass of PYL Proteins
Mol.Cell, 42, 2011
|
|
1ZZ5
 
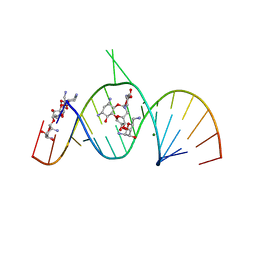 | | Molecular Recognition of RNA by Neomycin and a Restricted Neomycin Derivative | | Descriptor: | 13,15-DIAMINO-2-(AMINOMETHYL)-3,4,9,12-TETRAHYDROXYHEXADECAHYDRO-2H-7,10-EPOXYPYRANO[2,3-B][1,10,4]BENZODIOXAZACYCLODODECIN-8-YL 2,6-DIAMINO-2,6-DIDEOXYHEXOPYRANOSIDE, 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*AP*CP*C)-3', 5'-R(*GP*UP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*CP*GP*G)-3', ... | | Authors: | Zhao, Q, Zhao, F, Blount, K.F, Han, Q, Tor, Y, Hermann, T. | | Deposit date: | 2005-06-13 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular recognition of RNA by neomycin and a restricted neomycin derivative
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
2A04
 
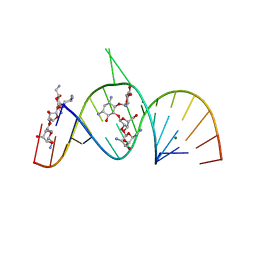 | | Molecular Recognition of RNA by Neomycin and a Restricted Neomycin Derivative | | Descriptor: | 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*AP*CP*C)-3', 5'-R(*GP*UP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*CP*GP*G)-3', MAGNESIUM ION, ... | | Authors: | Zhao, Q, Zhao, F, Blount, K.F, Han, Q, Tor, Y, Hermann, T. | | Deposit date: | 2005-06-15 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Molecular recognition of RNA by neomycin and a restricted neomycin derivative
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
2PN3
 
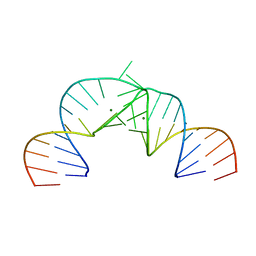 | | Crystal Structure of Hepatitis C Virus IRES Subdomain IIa | | Descriptor: | 5'-R(*CP*GP*GP*AP*GP*GP*AP*AP*CP*UP*AP*CP*UP*GP*UP*CP*UP*UP*CP*AP*CP*GP*CP*C)-3', 5'-R(*GP*CP*GP*(5BU)P*GP*UP*CP*GP*UP*GP*CP*AP*GP*CP*CP*(5BU)P*CP*CP*GP*G)-3', MAGNESIUM ION | | Authors: | Zhao, Q, Han, Q, Kissinger, C.R, Hermann, T, Thompson, P.A. | | Deposit date: | 2007-04-23 | | Release date: | 2008-04-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of hepatitis C virus IRES subdomain IIa.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
2PN4
 
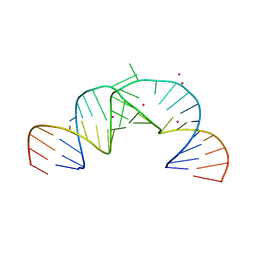 | | Crystal Structure of Hepatitis C Virus IRES Subdomain IIa | | Descriptor: | 5'-R(*CP*GP*GP*AP*GP*GP*AP*AP*CP*UP*AP*CP*UP*GP*UP*CP*UP*UP*CP*AP*CP*GP*CP*C)-3', 5'-R(*GP*CP*GP*(5BU)P*GP*UP*CP*GP*UP*GP*CP*AP*GP*CP*CP*(5BU)P*CP*CP*GP*G)-3', STRONTIUM ION | | Authors: | Zhao, Q, Han, Q, Kissinger, C.R, Thompson, P.A. | | Deposit date: | 2007-04-23 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structure of hepatitis C virus IRES subdomain IIa.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
3BR9
 
 | | Crystal Structure of HCV NS5B Polymerase with a Novel Pyridazinone Inhibitor | | Descriptor: | (2R)-2-({3-[5-hydroxy-2-(3-methylbutyl)-3-oxo-6-thiophen-2-yl-2,3-dihydropyridazin-4-yl]-1,1-dioxido-2H-1,2,4-benzothia diazin-7-yl}oxy)propanamide, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2007-12-21 | | Release date: | 2008-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Novel HCV NS5B polymerase inhibitors derived from 4-(1',1'-dioxo-1',4'-dihydro-1'lambda6-benzo[1',2',4']thiadiazin-3'-yl)-5-hydroxy-2H-pyridazin-3-ones. Part 1: exploration of 7'-substitution of benzothiadiazine.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3BSC
 
 | | Crystal Structure of HCV NS5B Polymerase with a Novel Pyridazinone Inhibitor | | Descriptor: | 5-hydroxy-4-(7-methoxy-1,1-dioxido-2H-1,2,4-benzothiadiazin-3-yl)-2-(3-methylbutyl)-6-phenylpyridazin-3(2H)-one, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2007-12-23 | | Release date: | 2008-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Novel HCV NS5B polymerase inhibitors derived from 4-(1',1'-dioxo-1',4'-dihydro-1'lambda6-benzo[1',2',4']thiadiazin-3'-yl)-5-hydroxy-2H-pyridazin-3-ones. Part 1: exploration of 7'-substitution of benzothiadiazine.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
8H2I
 
 | | Near-atomic structure of five-fold averaged PBCV-1 capsid | | Descriptor: | MCPv1, MCPv2, MCPv3, ... | | Authors: | Shao, Q, Agarkova, I.V, Noel, E.A, Dunigan, D.D, Liu, Y, Wang, A, Guo, M, Xie, L, Zhao, X, Rossmann, M.G, Van Etten, J.L, Klose, T, Fang, Q. | | Deposit date: | 2022-10-06 | | Release date: | 2022-11-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Near-atomic, non-icosahedrally averaged structure of giant virus Paramecium bursaria chlorella virus 1.
Nat Commun, 13, 2022
|
|
1ZX7
 
 | | Molecular Recognition of RNA by Neomycin and a Restricted Neomycin Derivative | | Descriptor: | 5'-R(*CP*GP*CP*GP*UP*CP*AP*CP*AP*CP*CP*AP*CP*C)-3', 5'-R(*GP*UP*GP*GP*UP*GP*AP*AP*GP*UP*CP*GP*CP*GP*G)-3' | | Authors: | Zhao, Q, Zhao, F, Blount, K.F, Han, Q, Tor, Y, Hermann, T. | | Deposit date: | 2005-06-07 | | Release date: | 2005-09-20 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Molecular recognition of RNA by neomycin and a restricted neomycin derivative
Angew.Chem.Int.Ed.Engl., 44, 2005
|
|
3GYN
 
 | | Crystal structure of HCV NS5B polymerase with a novel monocyclic dihydropyridinone inhibitor | | Descriptor: | N-{3-[(5R)-1-cyclopentyl-4-hydroxy-5-methyl-5-(3-methylbutyl)-2-oxo-1,2,5,6-tetrahydropyridin-3-yl]-1,1-dioxido-4H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2009-04-04 | | Release date: | 2009-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | 5,5'- and 6,6'-dialkyl-5,6-dihydro-1H-pyridin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3IGV
 
 | | Crystal structure of HCV NS5B polymerase with a novel monocyclic dihydro-pyridinone inhibitor | | Descriptor: | N-{3-[(6S)-6-ethyl-1-(4-fluorobenzyl)-4-hydroxy-2-oxo-1,2,5,6-tetrahydropyridin-3-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-DIRECTED RNA POLYMERASE | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2009-07-28 | | Release date: | 2009-12-08 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | 5,5'- and 6,6'-dialkyl-5,6-dihydro-1H-pyridin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3CDE
 
 | | Crystal structure of HCV NS5B polymerase with a novel Pyridazinone inhibitor | | Descriptor: | N-{3-[5-hydroxy-2-(3-methylbutyl)-3-oxo-6-thiophen-2-yl-2,3-dihydropyridazin-4-yl]-1,1-dioxido-2H-1,2,4-benzothiadiazin-7-yl}methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2008-02-26 | | Release date: | 2009-03-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Novel HCV NS5B polymerase inhibitors derived from 4-(1',1'-dioxo-1',4'-dihydro-1'lambda(6)-benzo[1',2',4']thiadiazin-3'-yl)-5-hydroxy-2H-pyridazin-3-ones. Part 3: Further optimization of the 2-, 6-, and 7'-substituents and initial pharmacokinetic assessments.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3D5M
 
 | | Crystal structure of HCV NS5B polymerase with a novel pyridazinone inhibitor | | Descriptor: | N-({3-[(5S)-5-tert-butyl-1-(3-chloro-4-fluorobenzyl)-4-hydroxy-2-oxo-2,5-dihydro-1H-pyrrol-3-yl]-1,1-dioxido-1,2-benzis othiazol-7-yl}methyl)methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2008-05-16 | | Release date: | 2009-05-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-based design, synthesis, and biological evaluation of 1,1-dioxoisothiazole and benzo[b]thiophene-1,1-dioxide derivatives as novel inhibitors of hepatitis C virus NS5B polymerase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1B1C
 
 | | CRYSTAL STRUCTURE OF THE FMN-BINDING DOMAIN OF HUMAN CYTOCHROME P450 REDUCTASE AT 1.93A RESOLUTION | | Descriptor: | CALCIUM ION, FLAVIN MONONUCLEOTIDE, PROTEIN (NADPH-CYTOCHROME P450 REDUCTASE) | | Authors: | Zhao, Q, Modi, S, Smith, G, Paine, M, Mcdonagh, P.D, Wolf, C.R, Tew, D, Lian, L.-Y, Roberts, G.C.K, Driessen, H.P.C. | | Deposit date: | 1998-11-19 | | Release date: | 1999-11-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of the FMN-binding domain of human cytochrome P450 reductase at 1.93 A resolution.
Protein Sci., 8, 1999
|
|
1A6Y
 
 | | REVERBA ORPHAN NUCLEAR RECEPTOR/DNA COMPLEX | | Descriptor: | DNA (5'-D(*CP*AP*AP*CP*TP*AP*GP*GP*TP*CP*AP*CP*(5IT)P*AP*GP*GP*TP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*AP*GP*TP*GP*AP*CP*CP*TP*AP*GP*TP*TP*G)-3'), ORPHAN NUCLEAR RECEPTOR NR1D1, ... | | Authors: | Zhao, Q, Khorasanizadeh, S, Rastinejad, F. | | Deposit date: | 1998-03-04 | | Release date: | 1998-10-21 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural elements of an orphan nuclear receptor-DNA complex.
Mol.Cell, 1, 1998
|
|
3CVK
 
 | | Crystal structure of HCV NS5B polymerase with a novel pyridazinone inhibitor | | Descriptor: | N-{3-[1-(3,3-Dimethyl-butyl)-4-hydroxy-2-oxo-1,2,4a,5,6,7-hexahydro-pyrrolo[1,2-b]pyridazin-3-yl]-1,1-dioxo-1,2-dihydro -1lambda6-benzo[1,2,4]thiadiazin-7-yl}-methanesulfonamide, RNA-directed RNA polymerase | | Authors: | Zhao, Q, Showalter, R.E, Han, Q, Kissinger, C.R. | | Deposit date: | 2008-04-18 | | Release date: | 2009-04-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Hexahydro-pyrrolo- and hexahydro-1H-pyrido[1,2-b]pyridazin-2-ones as potent inhibitors of HCV NS5B polymerase.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
1BY4
 
 | | STRUCTURE AND MECHANISM OF THE HOMODIMERIC ASSEMBLY OF THE RXR ON DNA | | Descriptor: | DNA (5'-D(*C*TP*AP*GP*GP*TP*CP*AP*AP*AP*GP*GP*TP*CP*AP*G)-3'), DNA (5'-D(*CP*TP*GP*AP*CP*CP*TP*TP*TP*GP*AP*CP*CP*TP*A)-3'), PROTEIN (RETINOIC ACID RECEPTOR RXR-ALPHA), ... | | Authors: | Zhao, Q, Chasse, S.A, Devarakonda, S, Sierk, M.L, Ahvazi, B, Sigler, P.B, Rastinejad, F. | | Deposit date: | 1998-10-22 | | Release date: | 2000-01-12 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of RXR-DNA interactions.
J.Mol.Biol., 296, 2000
|
|
1YDU
 
 | | Solution NMR structure of At5g01610, an Arabidopsis thaliana protein containing DUF538 domain | | Descriptor: | At5g01610 | | Authors: | Zhao, Q, Cornilescu, C.C, Lee, M.S, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2004-12-26 | | Release date: | 2005-02-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of At5g01610, an Arabidopsis thaliana protein containing DUF538 domain
To be Published
|
|
4REB
 
 | | Structural Insights into 5' Flap DNA Unwinding and Incision by the Human FAN1 Dimer | | Descriptor: | DNA (5'-D(P*CP*GP*TP*GP*GP*CP*GP*AP*GP*CP*GP*CP*TP*CP*GP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*GP*CP*TP*CP*GP*CP*CP*AP*CP*G)-3'), DNA (5'-D(P*GP*TP*GP*GP*CP*GP*AP*GP*C)-3'), ... | | Authors: | Zhao, Q, Xue, X, Longerich, S, Sung, P, Xiong, Y. | | Deposit date: | 2014-09-22 | | Release date: | 2014-12-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structural insights into 5' flap DNA unwinding and incision by the human FAN1 dimer.
Nat Commun, 5, 2014
|
|
4REC
 
 | | A nuclease-DNA complex form 3 | | Descriptor: | DNA (40-MER), Fanconi-associated nuclease 1, IODIDE ION | | Authors: | Zhao, Q, Xue, X, Longerich, S, Sung, P, Xiong, Y. | | Deposit date: | 2014-09-22 | | Release date: | 2014-12-24 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural insights into 5' flap DNA unwinding and incision by the human FAN1 dimer.
Nat Commun, 5, 2014
|
|
