2H1W
 
 | | Crystal structure of the His183Ala mutant variant of Bacillus subtilis ferrochelatase | | Descriptor: | FE (II) ION, Ferrochelatase, MAGNESIUM ION | | Authors: | Hansson, M.D, Karlberg, T, Arys Rahardja, M, Al-Karadaghi, S, Hansson, M. | | Deposit date: | 2006-05-17 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Amino Acid Residues His183 and Glu264 in Bacillus subtilis Ferrochelatase Direct and Facilitate the Insertion of Metal Ion into Protoporphyrin IX
Biochemistry, 46, 2007
|
|
2H1V
 
 | | Crystal structure of the Lys87Ala mutant variant of Bacillus subtilis ferrochelatase | | Descriptor: | Ferrochelatase, MAGNESIUM ION | | Authors: | Hansson, M.D, Karlberg, T, Arys Rahardja, M, Al-Karadaghi, S, Hansson, M. | | Deposit date: | 2006-05-17 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Amino Acid Residues His183 and Glu264 in Bacillus subtilis Ferrochelatase Direct and Facilitate the Insertion of Metal Ion into Protoporphyrin IX
Biochemistry, 46, 2007
|
|
1PRB
 
 | | STRUCTURE OF AN ALBUMIN-BINDING DOMAIN, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | PROTEIN PAB | | Authors: | Johansson, M.U, De Chateau, M, Wikstrom, M, Forsen, S, Drakenberg, T, Bjorck, L. | | Deposit date: | 1997-01-15 | | Release date: | 1997-07-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the albumin-binding GA module: a versatile bacterial protein domain.
J.Mol.Biol., 266, 1997
|
|
1W7W
 
 | |
1GJS
 
 | | Solution structure of the Albumin binding domain of Streptococcal Protein G | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN G | | Authors: | Johansson, M.U, Frick, I.M, Nilsson, H, Kraulis, P.J, Hober, S, Jonasson, P, Nygren, A.P, Uhlen, M, Bjorck, L, Drakenberg, T, Forsen, S, Wikstrom, M. | | Deposit date: | 2001-08-02 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Specificity, and Mode of Interaction for Bacterial Albumin-Binding Modules
J.Biol.Chem., 277, 2002
|
|
1GAB
 
 | | STRUCTURE OF AN ALBUMIN-BINDING DOMAIN, NMR, 20 STRUCTURES | | Descriptor: | PROTEIN PAB | | Authors: | Johansson, M.U, De Chateau, M, Wikstrom, M, Forsen, S, Drakenberg, T, Bjorck, L. | | Deposit date: | 1996-12-30 | | Release date: | 1997-07-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the albumin-binding GA module: a versatile bacterial protein domain.
J.Mol.Biol., 266, 1997
|
|
1GJT
 
 | | Solution structure of the Albumin binding domain of Streptococcal Protein G | | Descriptor: | IMMUNOGLOBULIN G BINDING PROTEIN G | | Authors: | Johansson, M.U, Frick, I.M, Nilsson, H, Kraulis, P.J, Hober, S, Jonasson, P, Nygren, A.P, Uhlen, M, Bjorck, L, Drakenberg, T, Forsen, S, Wikstrom, M. | | Deposit date: | 2001-08-02 | | Release date: | 2001-08-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure, Specificity, and Mode of Interaction for Bacterial Albumin-Binding Modules
J.Biol.Chem., 277, 2002
|
|
2JMM
 
 | | NMR solution structure of a minimal transmembrane beta-barrel platform protein | | Descriptor: | Outer membrane protein A | | Authors: | Johansson, M.U, Alioth, S, Hu, K, Walser, R, Koebnik, R, Pervushin, K. | | Deposit date: | 2006-11-20 | | Release date: | 2007-07-03 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | A minimal transmembrane beta-barrel platform protein studied by nuclear magnetic resonance
Biochemistry, 46, 2007
|
|
1LD3
 
 | | Crystal Structure of B. subilis ferrochelatase with Zn(2+) bound at the active site. | | Descriptor: | Ferrochelatase, ZINC ION | | Authors: | Lecerof, D, Fodje, M.N, Leon, R.A, Olsson, U, Hansson, A, Sigfridsson, E, Ryde, U, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 2002-04-08 | | Release date: | 2003-05-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Metal binding to Bacillus subtilis ferrochelatase and interaction between metal sites
J.Biol.Inorg.Chem., 8, 2003
|
|
3GOQ
 
 | |
1N0I
 
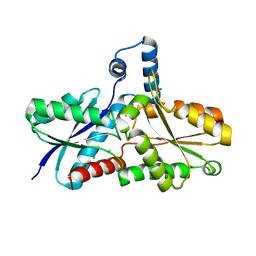 | | Crystal Structure of Ferrochelatase with Cadmium bound at active site | | Descriptor: | CADMIUM ION, CHLORIDE ION, Ferrochelatase, ... | | Authors: | Lecerof, D, Fodje, M.N, Leon, R.A, Olsson, U, Hansson, A, Sigfridsson, E, Ryde, U, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 2002-10-14 | | Release date: | 2003-05-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal binding to Bacillus subtilis ferrochelatase and interaction between metal sites
J.Biol.Inorg.Chem., 8, 2003
|
|
1G8P
 
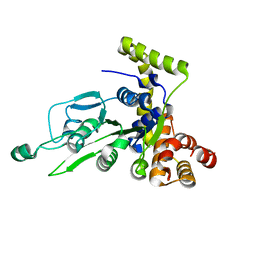 | | CRYSTAL STRUCTURE OF BCHI SUBUNIT OF MAGNESIUM CHELATASE | | Descriptor: | MAGNESIUM-CHELATASE 38 KDA SUBUNIT | | Authors: | Fodje, M.N, Hansson, A, Hansson, M, Olsen, J.G, Gough, S, Willows, R.D, Al-Karadaghi, S. | | Deposit date: | 2000-11-20 | | Release date: | 2001-08-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Interplay between an AAA module and an integrin I domain may regulate the function of magnesium chelatase.
J.Mol.Biol., 311, 2001
|
|
3M4Z
 
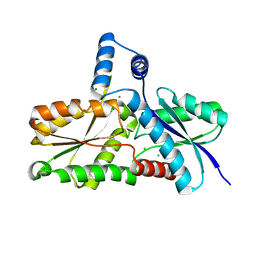 | | Crystal Structure of B. subtilis ferrochelatase with Cobalt bound at the active site | | Descriptor: | CHLORIDE ION, COBALT (II) ION, Ferrochelatase, ... | | Authors: | Soderberg, C.A.G, Hansson, M.D, Sreekanth, R, Al-Karadaghi, S, Hansson, M. | | Deposit date: | 2010-03-12 | | Release date: | 2010-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Bacterial ferrochelatase goes human: Tyr13 determines the apparent metal specificity of Bacillus subtilis ferrochelatase
To be Published
|
|
1DOZ
 
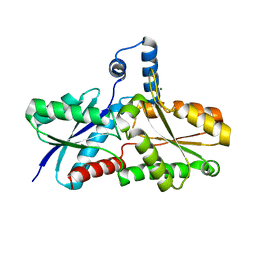 | | CRYSTAL STRUCTURE OF FERROCHELATASE | | Descriptor: | FERROCHELATASE, MAGNESIUM ION | | Authors: | Lecerof, D, Fodje, M, Hansson, A, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 1999-12-22 | | Release date: | 2000-01-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural and mechanistic basis of porphyrin metallation by ferrochelatase.
J.Mol.Biol., 297, 2000
|
|
1C1H
 
 | | CRYSTAL STRUCTURE OF BACILLUS SUBTILIS FERROCHELATASE IN COMPLEX WITH N-METHYL MESOPORPHYRIN | | Descriptor: | FERROCHELATASE, MAGNESIUM ION, N-METHYLMESOPORPHYRIN | | Authors: | Lecerof, D, Fodje, M, Hansson, A, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 1999-07-22 | | Release date: | 2000-03-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and mechanistic basis of porphyrin metallation by ferrochelatase.
J.Mol.Biol., 297, 2000
|
|
1C9E
 
 | | STRUCTURE OF FERROCHELATASE WITH COPPER(II) N-METHYLMESOPORPHYRIN COMPLEX BOUND AT THE ACTIVE SITE | | Descriptor: | MAGNESIUM ION, N-METHYLMESOPORPHYRIN CONTAINING COPPER, PROTOHEME FERROLYASE | | Authors: | Lecerof, D, Fodje, M.N, Hansson, A, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 1999-08-02 | | Release date: | 2000-03-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and mechanistic basis of porphyrin metallation by ferrochelatase.
J.Mol.Biol., 297, 2000
|
|
2AC2
 
 | | Crystal structure of the Tyr13Phe mutant variant of Bacillus subtilis Ferrochelatase with Zn(2+) bound at the active site | | Descriptor: | Ferrochelatase, ZINC ION | | Authors: | Shipovskov, S, Karlberg, T, Fodje, M, Hansson, M.D, Ferreira, G.C, Hansson, M, Reimann, C.T, Al-Karadaghi, S. | | Deposit date: | 2005-07-18 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Metallation of the Transition-state Inhibitor N-methyl Mesoporphyrin by Ferrochelatase: Implications for the Catalytic Reaction Mechanism.
J.Mol.Biol., 352, 2005
|
|
2AC4
 
 | | Crystal structure of the His183Cys mutant variant of Bacillus subtilis Ferrochelatase | | Descriptor: | Ferrochelatase | | Authors: | Shipovskov, S, Karlberg, T, Fodje, M, Hansson, M.D, Ferreira, G.C, Hansson, M, Reimann, C.T, Al-Karadaghi, S. | | Deposit date: | 2005-07-18 | | Release date: | 2005-09-20 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Metallation of the Transition-state Inhibitor N-methyl Mesoporphyrin by Ferrochelatase: Implications for the Catalytic Reaction Mechanism.
J.Mol.Biol., 352, 2005
|
|
8S03
 
 | | NMR solution structure of the CysD2 domain of MUC2 | | Descriptor: | CALCIUM ION, Mucin-2 | | Authors: | Recktenwald, C, Karlsson, B.G, Garcia-Bonnete, M.-J, Katona, G, Jensen, M, Lymer, R, Baeckstroem, M, Johansson, M.E.V, Hansson, G.C, Trillo-Muyo, S. | | Deposit date: | 2024-02-13 | | Release date: | 2024-04-17 | | Last modified: | 2024-11-06 | | Method: | SOLUTION NMR | | Cite: | The structure of the second CysD domain of MUC2 and role in mucin organization by transglutaminase-based cross-linking.
Cell Rep, 43, 2024
|
|
8RDE
 
 | |
8R0T
 
 | |
1LBQ
 
 | | The crystal structure of Saccharomyces cerevisiae ferrochelatase | | Descriptor: | Ferrochelatase | | Authors: | Karlberg, T, Lecerof, D, Gora, M, Silvegren, G, Labbe-Bois, R, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 2002-04-04 | | Release date: | 2002-11-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Metal binding to Saccharomyces cerevisiae ferrochelatase
Biochemistry, 41, 2002
|
|
1L8X
 
 | | Crystal Structure of Ferrochelatase from the Yeast, Saccharomyces cerevisiae, with Cobalt(II) as the Substrate Ion | | Descriptor: | COBALT (II) ION, Ferrochelatase | | Authors: | Karlberg, T, Lecerof, D, Gora, M, Silvegren, G, Labbe-Bois, R, Hansson, M, Al-Karadaghi, S. | | Deposit date: | 2002-03-22 | | Release date: | 2002-11-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Metal Binding to Saccharomyces cerevisiae Ferrochelatase
Biochemistry, 41, 2002
|
|
1AK1
 
 | | FERROCHELATASE FROM BACILLUS SUBTILIS | | Descriptor: | FERROCHELATASE | | Authors: | Al-Karadaghi, S, Hansson, M, Nikonov, S, Jonsson, B, Hederstedt, L. | | Deposit date: | 1997-05-28 | | Release date: | 1997-12-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of ferrochelatase: the terminal enzyme in heme biosynthesis.
Structure, 5, 1997
|
|
2X31
 
 | | Modelling of the complex between subunits BchI and BchD of magnesium chelatase based on single-particle cryo-EM reconstruction at 7.5 ang | | Descriptor: | MAGNESIUM-CHELATASE 38 KDA SUBUNIT, MAGNESIUM-CHELATASE 60 KDA SUBUNIT | | Authors: | Lunqvist, J, Elmlund, H, Peterson Wulff, R, Berglund, L, Elmlund, D, Emanuelsson, C, Hebert, H, Willows, R.D, Hansson, M, Lindahl, M, Al-Karadaghi, S. | | Deposit date: | 2010-01-19 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.5 Å) | | Cite: | ATP-Induced Conformational Dynamics in the Aaa+ Motor Unit of Magnesium Chelatase.
Structure, 18, 2010
|
|
