2M71
 
 | | Solution structure of the a C-terminal domain of translation initiation factor IF-3 from Campylobacter jejuni | | Descriptor: | Translation initiation factor IF-3 | | Authors: | Harris, R, Ahmed, M, Attonito, J, Bonanno, J.B, Chamala, S, Chowdhury, S, Evans, B, Fiser, A, Glenn, A.S, Hammonds, J, Hillerich, B, Khafizov, K, Lafleur, J, Love, J.D, Seidel, R.D, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-16 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the a C-terminal domain of translation initiation factor IF-3 from Campylobacter jejuni
To be Published
|
|
2M72
 
 | | Solution structure of uncharacterized thioredoxin-like protein PG_2175 from Porphyromonas gingivalis | | Descriptor: | Uncharacterized thioredoxin-like protein | | Authors: | Harris, R, Ahmed, M, Attonito, J, Bonanno, J.B, Chamala, S, Chowdhury, S, Evans, B, Fiser, A, Glenn, A.S, Hammonds, J, Hillerich, B, Khafizov, K, Lafleur, J, Love, J.D, Seidel, R.D, Stead, M, Girvin, M.E, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-04-16 | | Release date: | 2013-05-15 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of uncharacterized thioredoxin-like protein PG_2175 from Porphyromonas gingivalis
To be Published
|
|
4WBT
 
 | | Crystal structure of histidinol-phosphate aminotransferase from Sinorhizobium meliloti in complex with pyridoxal-5'-phosphate | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Shabalin, I.G, Bacal, P, Kowalska, A.K, Cooper, D.R, Stead, M, Hammonds, J, Ahmed, M, Hillerich, B.S, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-03 | | Release date: | 2014-09-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of histidinol-phosphate aminotransferase from Sinorhizobium meliloti in complex with pyridoxal-5'-phosphate
to be published
|
|
4WGH
 
 | | Crystal structure of aldo/keto reductase from Klebsiella pneumoniae in complex with NADP and acetate at 1.8 A resolution | | Descriptor: | ACETATE ION, Aldehyde reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Bacal, P, Shabalin, I.G, Cooper, D.R, Hillerich, B.S, Zimmerman, M.D, Chowdhury, S, Hammonds, J, Al Obaidi, N, Gizzi, A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-09-18 | | Release date: | 2014-10-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of aldo/keto reductase from Klebsiella pneumoniae in complex with NADP and acetate at 1.8 A resolution
to be published
|
|
4NHZ
 
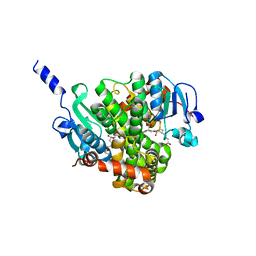 | | Crystal structure of glutathione transferase BBTA-3750 from Bradyrhizobium sp., Target EFI-507290, with one glutathione bound | | Descriptor: | GLUTATHIONE, Putative glutathione S-transferase enzyme with thioredoxin-like domain | | Authors: | Patskovsky, Y, Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-05 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Crystal Structure of Glutathione Transferase Bbta-3750 from Bradyrhizobium Sp., Target Efi-507290
To be Published
|
|
4NHW
 
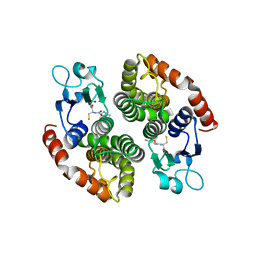 | | Crystal structure of glutathione transferase SMc00097 from Sinorhizobium meliloti, target EFI-507275, with one glutathione bound per one protein subunit | | Descriptor: | GLUTATHIONE, Glutathione S-transferase | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-11-05 | | Release date: | 2013-11-20 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of glutathione transferase SMc00097 from Sinorhizobium meliloti, target EFI-507275
To be Published
|
|
4Q60
 
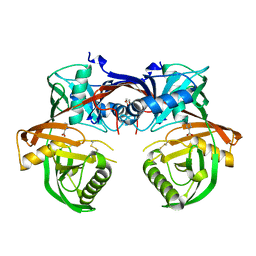 | | Crystal structure of a 4-hydroxyproline epimerase from Burkholderia Multivorans atcc 17616, target EFI-506586, open form, with bound pyrrole-2-carboxylate | | Descriptor: | GLYCEROL, PROLINE RACEMASE, PYRROLE-2-CARBOXYLATE | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N, Sojitra, S, Stead, M, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-18 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | CRYSTAL STRUCTURE OF PROLINE RACEMASE Bmul_4447 FROM Burkholderia multivorans, TARGET EFI-506586
To be Published
|
|
4MPG
 
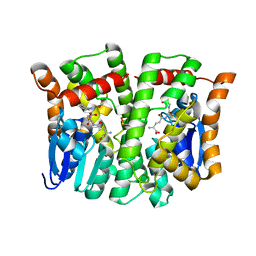 | | Crystal structure of human glutathione transferase theta-2, complex with glutathione and unknown ligand, target EFI-507257 | | Descriptor: | FORMIC ACID, GLUTATHIONE, Glutathione S-transferase theta-2, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-12 | | Release date: | 2013-09-25 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.951 Å) | | Cite: | Crystal Structure of Human Glutathione S-Transferase Theta-2 (Target EFI-507257)
To be Published
|
|
4PX1
 
 | | CRYSTAL STRUCTURE OF Maleylacetoacetate isomerase from Methylobacteriu extorquens AM1 WITH BOUND MALONATE (TARGET EFI-507068) | | Descriptor: | CHLORIDE ION, MALONIC ACID, Maleylacetoacetate isomerase (Glutathione S-transferase) | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-21 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of glutathione s-transferase zeta from methylobacterium extorquens (TARGET EFI-507068)
To be Published
|
|
4MPF
 
 | | Crystal structure of human glutathione transferase theta-2, complex with inorganic phosphate, GSH free, target EFI-507257 | | Descriptor: | Glutathione S-transferase theta-2, L(+)-TARTARIC ACID, PHOSPHATE ION | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-12 | | Release date: | 2013-09-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Human Glutathione S-Transferase Theta-2 (Target Efi-507257)
To be Published
|
|
4NAX
 
 | | Crystal structure of glutathione transferase PPUT_1760 from Pseudomonas putida, target EFI-507288, with one glutathione disulfide bound per one protein subunit | | Descriptor: | FORMIC ACID, GLYCEROL, Glutathione S-transferase, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-22 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.301 Å) | | Cite: | Crystal structure of glutathione transferase Pput_1760 from Pseudomonas putida, target EFI-507288
To be Published
|
|
4MZW
 
 | | CRYSTAL STRUCTURE OF NU-CLASS GLUTATHIONE TRANSFERASE YGHU FROM Streptococcus sanguinis SK36, COMPLEX WITH GLUTATHIONE DISULFIDE, TARGET EFI-507286 | | Descriptor: | ACETATE ION, Glutathione S-Transferase, OXIDIZED GLUTATHIONE DISULFIDE | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-09-30 | | Release date: | 2013-10-16 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase Yghu (Target Efi-507286)
To be Published
|
|
4PXO
 
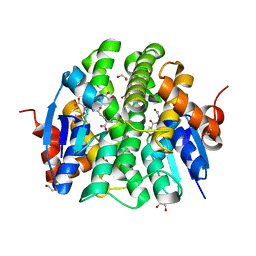 | | Crystal structure of Maleylacetoacetate isomerase from Methylobacteriu extorquens AM1 WITH BOUND MALONATE AND GSH (TARGET EFI-507068) | | Descriptor: | 1,2-ETHANEDIOL, GLUTATHIONE, MALONIC ACID, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Al Obaidi, N, Stead, M, Love, J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-03-24 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of glutathione s-transferase zeta from Methylobacterium extorquens (TARGET EFI-507068)
To be Published
|
|
4Q2H
 
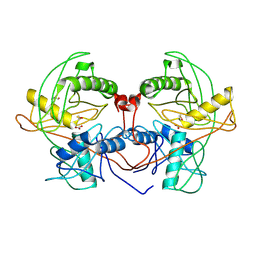 | | Crystal structure of probable proline racemase from agrobacterium radiobacter K84, TARGET EFI-506561, with bound carbonate | | Descriptor: | BICARBONATE ION, GLYCEROL, Proline racemase protein | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Glenn, A.S, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-04-08 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Proline Racemase Arad_0731 from Agrobacterium Radiobacter, Target Efi-506561
To be Published
|
|
4O9K
 
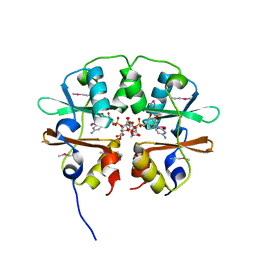 | | Crystal structure of the CBS pair of a putative D-arabinose 5-phosphate isomerase from Methylococcus capsulatus in complex with CMP-Kdo | | Descriptor: | Arabinose 5-phosphate isomerase, CYTIDINE 5'-MONOPHOSPHATE 3-DEOXY-BETA-D-GULO-OCT-2-ULO-PYRANOSONIC ACID, GLYCEROL | | Authors: | Shabalin, I.G, Cooper, D.R, Shumilin, I.A, Zimmerman, M.D, Majorek, K.A, Hammonds, J, Hillerich, B.S, Nawar, A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-01-02 | | Release date: | 2014-01-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure and kinetic properties of D-arabinose 5-phosphate isomerase from Methylococcus capsulatus
To be Published
|
|
4XCV
 
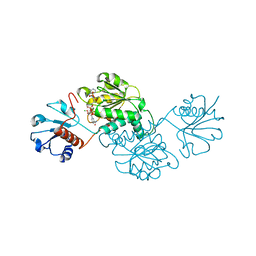 | | Probable 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADPH | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, NADP-dependent 2-hydroxyacid dehydrogenase, ... | | Authors: | Langner, K.M, Shabalin, I.G, Handing, K.B, Gasiorowska, O.A, Stead, M, Hillerich, B.S, Chowdhury, S, Hammonds, J, Zimmerman, M.D, Al Obadi, N, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-12-18 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of 2-hydroxyacid dehydrogenase from Rhizobium etli CFN 42 in complex with NADPH
to be published
|
|
4YYC
 
 | | Crystal structure of trimethylamine methyltransferase from Sinorhizobium meliloti in complex with unknown ligand | | Descriptor: | CHLORIDE ION, Putative trimethylamine methyltransferase, UNKNOWN LIGAND | | Authors: | Shabalin, I.G, Porebski, P.J, Gasiorowska, O.A, Handing, K.B, Niedzialkowska, E, Cymborowski, M.T, Cooper, D.R, Stead, M, Hammonds, J, Ahmed, M, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-03-23 | | Release date: | 2015-04-08 | | Last modified: | 2022-04-13 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Protein purification and crystallization artifacts: The tale usually not told.
Protein Sci., 25, 2016
|
|
4N18
 
 | | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase family protein from Klebsiella pneumoniae 342 | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CITRIC ACID, D-isomer specific 2-hydroxyacid dehydrogenase family protein | | Authors: | Bacal, P, Shabalin, I.G, Cooper, D.R, Majorek, K.A, Osinski, T, Hillerich, B.S, Hammonds, J, Nawar, A, Stead, M, Chowdhury, S, Gizzi, A, Bonanno, J, Seidel, R, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-10-03 | | Release date: | 2013-10-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Crystal structure of D-isomer specific 2-hydroxyacid dehydrogenase family protein from Klebsiella pneumoniae 342
To be Published
|
|
3TWA
 
 | | Crystal structure of gluconate dehydratase (TARGET EFI-501679) from Salmonella enterica subsp. enterica serovar Enteritidis str. P125109 complexed with magnesium and glycerol | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-09-21 | | Release date: | 2011-10-26 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Gluconate Dehydratase from Salmonella Enterica P125109
To be Published
|
|
3T6C
 
 | | Crystal structure of an enolase from pantoea ananatis (efi target efi-501676) with bound d-gluconate and mg | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, D-gluconic acid, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-07-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.598 Å) | | Cite: | Crystal structure of an enolase from pantoea ananatis (efi target efi-501676) with bound d-gluconate and mg
to be published
|
|
3THU
 
 | | Crystal structure of an enolase from sphingomonas sp. ska58 (efi target efi-501683) with bound mg | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Wasserman, S.R, Morisco, L.L, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-08-19 | | Release date: | 2011-09-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of an enolase from sphingomonas sp. ska58 (efi target efi-501683) with bound mg
to be published
|
|
3V7P
 
 | | Crystal structure of amidohydrolase nis_0429 (target efi-500396) from Nitratiruptor sp. sb155-2 | | Descriptor: | Amidohydrolase family protein, BENZOIC ACID, BICARBONATE ION, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Raushel, F.M, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-12-21 | | Release date: | 2012-01-11 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of Amidohydrolase Nis_0429 (Target Efi-500319) from Nitratiruptor Sp. Sb155-2
To be Published
|
|
3UJ2
 
 | | CRYSTAL STRUCTURE OF AN ENOLASE FROM ANAEROSTIPES CACCAE (EFI TARGET EFI-502054) WITH BOUND MG AND SULFATE | | Descriptor: | Enolase 1, MAGNESIUM ION, SULFATE ION | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Hillerich, B, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-11-07 | | Release date: | 2011-11-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF AN ENOLASE FROM ANAEROSTIPES CACCAE (EFI TARGET EFI-502054) WITH BOUND MG AND SULFATE
to be published
|
|
3VCN
 
 | | Crystal structure of mannonate dehydratase (target EFI-502209) from Caulobacter crescentus CB15 | | Descriptor: | CARBONATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Zencheck, W.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2012-01-04 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of mannonate dehydratase from Caulobacter crescentus CB15
To be Published
|
|
3UBK
 
 | | Crystal structure of glutathione transferase (TARGET EFI-501770) from leptospira interrogans | | Descriptor: | CHLORIDE ION, GLYCEROL, Glutathione transferase, ... | | Authors: | Patskovsky, Y, Toro, R, Bhosle, R, Zencheck, W.D, Hillerich, B, Seidel, R.D, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Imker, H.J, Armstrong, R.N, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2011-10-24 | | Release date: | 2011-11-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of Glutathione S-Transferase from Leptospira Interrogans
To be Published
|
|
