8RTW
 
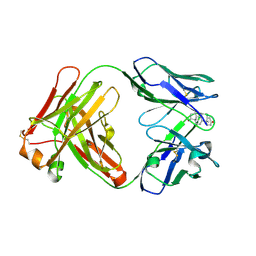 | | Crystal Structure of the Anti-testosterone Fab in Complex with Testosterone | | Descriptor: | Anti-testosterone Fab 220 heavy chain, Anti-testosterone Fab 220 light chain, TESTOSTERONE | | Authors: | Eronen, V, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2024-01-29 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural insights into ternary immunocomplex formation and cross-reactivity: binding of an anti-immunocomplex FabB12 to Fab220-testosterone complex.
Febs J., 2024
|
|
8RTX
 
 | |
1XNK
 
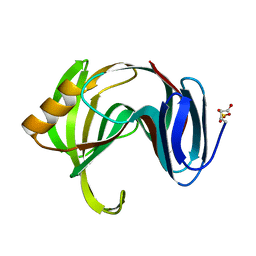 | | Beta-1,4-xylanase from Chaetomium thermophilum complexed with methyl thioxylopentoside | | Descriptor: | 4-thio-beta-D-xylopyranose-(1-4)-4-thio-beta-D-xylopyranose-(1-4)-methyl 4-thio-alpha-D-xylopyranoside, SULFATE ION, endoxylanase 11A | | Authors: | Hakanpaa, J, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2004-10-05 | | Release date: | 2005-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Determination of thioxylo-oligosaccharide binding to family 11 xylanases using electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry and X-ray crystallography
FEBS J., 272, 2005
|
|
8CBY
 
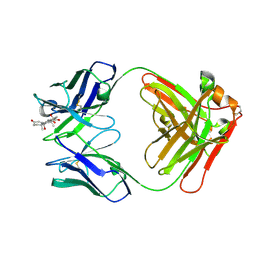 | | Crystal Structure of Anti-cortisol Fab in Complex with Cortisol | | Descriptor: | (11alpha,14beta)-11,17,21-trihydroxypregn-4-ene-3,20-dione, anti-cortisol (17) Fab (heavy chain), anti-cortisol (17) Fab (light chain) | | Authors: | Eronen, V, Rouvinen, J, Hakulinen, N. | | Deposit date: | 2023-01-26 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural insight to elucidate the binding specificity of the anti-cortisol Fab fragment with glucocorticoids.
J.Struct.Biol., 215, 2023
|
|
8CBX
 
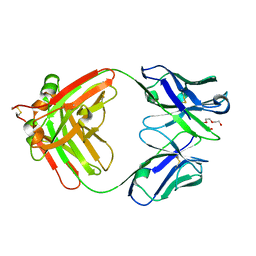 | | Crystal Structure of Anti-Cortisol Fab fragment | | Descriptor: | TETRAETHYLENE GLYCOL, anti-cortisol (17) Fab (heavy chain), anti-cortisol (17) Fab (light chain) | | Authors: | Eronen, V, Rouvinen, J, Hakulinen, N. | | Deposit date: | 2023-01-26 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight to elucidate the binding specificity of the anti-cortisol Fab fragment with glucocorticoids.
J.Struct.Biol., 215, 2023
|
|
8CBZ
 
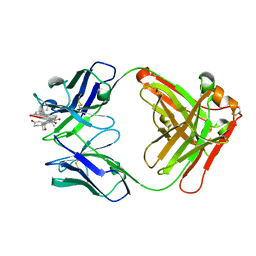 | |
6GSG
 
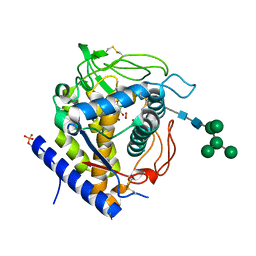 | | Crystal structure of Aspergillus oryzae catechol oxidase complexed with resorcinol | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, Catechol oxidase, ... | | Authors: | Penttinen, L, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2018-06-14 | | Release date: | 2018-09-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.192 Å) | | Cite: | Unraveling Substrate Specificity and Catalytic Promiscuity of Aspergillus oryzae Catechol Oxidase.
Chembiochem, 19, 2018
|
|
6KVE
 
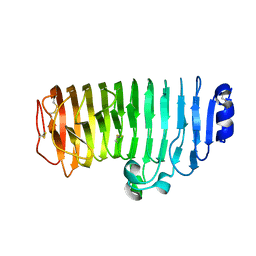 | |
8QPS
 
 | |
2R56
 
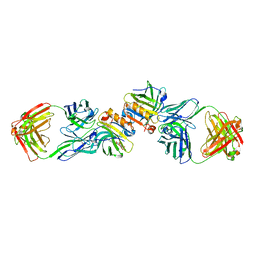 | | Crystal Structure of a Recombinant IgE Fab Fragment in Complex with Bovine Beta-Lactoglobulin Allergen | | Descriptor: | Beta-lactoglobulin, DODECYL-BETA-D-MALTOSIDE, IgE Fab Fragment, ... | | Authors: | Niemi, M, Kallio, J.M, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2007-09-03 | | Release date: | 2007-11-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular interactions between a recombinant IgE antibody and the beta-lactoglobulin allergen.
Structure, 15, 2007
|
|
5HWM
 
 | | Crystal structure of keto-deoxy-D-galactarate dehydratase complexed with 2-oxoadipic acid | | Descriptor: | 2-OXOADIPIC ACID, FORMIC ACID, Probable 5-dehydro-4-deoxyglucarate dehydratase | | Authors: | Taberman, H, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-23 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structure and function of a decarboxylating Agrobacterium tumefaciens keto-deoxy-d-galactarate dehydratase.
Biochemistry, 53, 2014
|
|
5HWJ
 
 | | Crystal structure of keto-deoxy-D-galactarate dehydratase | | Descriptor: | FORMIC ACID, GLYCEROL, Probable 5-dehydro-4-deoxyglucarate dehydratase | | Authors: | Taberman, H, Parkkinen, T, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2016-01-29 | | Release date: | 2016-03-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Structure and function of a decarboxylating Agrobacterium tumefaciens keto-deoxy-d-galactarate dehydratase.
Biochemistry, 53, 2014
|
|
7PLD
 
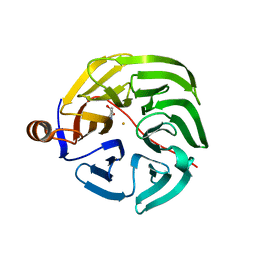 | |
7PLB
 
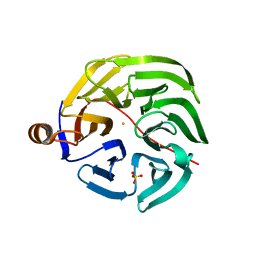 | | Caulobacter crescentus xylonolactonase with D-xylose | | Descriptor: | FE (II) ION, SULFATE ION, Smp-30/Cgr1 family protein, ... | | Authors: | Paakkonen, J, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2021-08-30 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Three-dimensional structure of xylonolactonase from Caulobacter crescentus: A mononuclear iron enzyme of the 6-bladed beta-propeller hydrolase family.
Protein Sci., 31, 2022
|
|
7PLC
 
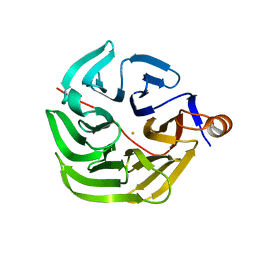 | | Caulobacter crescentus xylonolactonase with D-xylose, P21 space group | | Descriptor: | FE (II) ION, SULFATE ION, Smp-30/Cgr1 family protein, ... | | Authors: | Paakkonen, J, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2021-08-30 | | Release date: | 2021-11-24 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Three-dimensional structure of xylonolactonase from Caulobacter crescentus: A mononuclear iron enzyme of the 6-bladed beta-propeller hydrolase family.
Protein Sci., 31, 2022
|
|
6Z9I
 
 | | Escherichia coli D-2-deoxyribose-5-phosphate aldolase - N21K mutant complex with reaction products | | Descriptor: | 1,2-ETHANEDIOL, Deoxyribose-phosphate aldolase, GLYCERALDEHYDE-3-PHOSPHATE, ... | | Authors: | Paakkonen, J, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Substrate specificity of 2-deoxy-D-ribose 5-phosphate aldolase (DERA) assessed by different protein engineering and machine learning methods.
Appl.Microbiol.Biotechnol., 104, 2020
|
|
6Z9J
 
 | |
6Z9H
 
 | | Escherichia coli D-2-deoxyribose-5-phosphate aldolase - C47V/G204A/S239D mutant | | Descriptor: | 1,2-ETHANEDIOL, Deoxyribose-phosphate aldolase, FORMIC ACID, ... | | Authors: | Paakkonen, J, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2020-06-04 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Substrate specificity of 2-deoxy-D-ribose 5-phosphate aldolase (DERA) assessed by different protein engineering and machine learning methods.
Appl.Microbiol.Biotechnol., 104, 2020
|
|
5J83
 
 | |
5J84
 
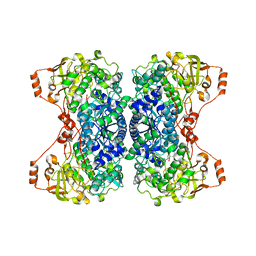 | |
5J85
 
 | | Ser480Ala mutant of L-arabinonate dehydratase | | Descriptor: | Dihydroxyacid dehydratase/phosphogluconate dehydratase, FE2/S2 (INORGANIC) CLUSTER, MAGNESIUM ION | | Authors: | Rahman, M.M, Rouvinen, J, Hakulinen, N. | | Deposit date: | 2016-04-07 | | Release date: | 2017-06-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Crystal Structure of a Bacterial l-Arabinonate Dehydratase Contains a [2Fe-2S] Cluster.
ACS Chem. Biol., 12, 2017
|
|
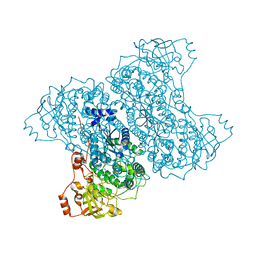
3PPS
 
 | | Crystal structure of an ascomycete fungal laccase from Thielavia arenaria | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, ... | | Authors: | Kallio, J.P, Rouvinen, J, Hakulinen, N. | | Deposit date: | 2010-11-25 | | Release date: | 2011-05-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an ascomycete fungal laccase from Thielavia arenaria--common structural features of asco-laccases.
Febs J., 278, 2011
|
|
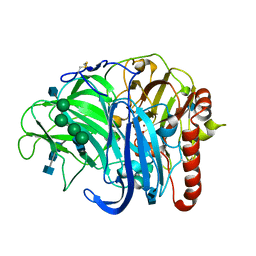
3QPK
 
 | | Probing oxygen channels in Melanocarpus albomyces laccase | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, ... | | Authors: | Kallio, J.P, Rouvinen, J, Hakulinen, N. | | Deposit date: | 2011-02-14 | | Release date: | 2011-05-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the Dioxygen Route in Melanocarpus albomyces Laccase with Pressurized Xenon Gas.
Biochemistry, 50, 2011
|
|
3TTY
 
 | | Crystal structure of beta-galactosidase from Bacillus circulans sp. alkalophilus in complex with galactose | | Descriptor: | Beta-galactosidase, ZINC ION, alpha-D-galactopyranose | | Authors: | Maksimainen, M, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2011-09-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural analysis, enzymatic characterization, and catalytic mechanisms of beta-galactosidase from Bacillus circulans sp. alkalophilus.
Febs J., 279, 2012
|
|
3TTS
 
 | | Crystal structure of beta-galactosidase from Bacillus circulans sp. alkalophilus | | Descriptor: | Beta-galactosidase, ZINC ION | | Authors: | Maksimainen, M, Hakulinen, N, Rouvinen, J. | | Deposit date: | 2011-09-15 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Structural analysis, enzymatic characterization, and catalytic mechanisms of beta-galactosidase from Bacillus circulans sp. alkalophilus.
Febs J., 279, 2012
|
|
