2BJW
 
 | | PspF AAA domain | | Descriptor: | PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Beuron, F, Niwa, H, Bordes, P, Wigneshweraraj, S, Keetch, C.A, Robinson, C.V, Buck, M, Zhang, X. | | Deposit date: | 2005-02-08 | | Release date: | 2005-03-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Insights Into the Activity of Enhancer-Binding Proteins
Science, 307, 2005
|
|
2BJV
 
 | | Crystal Structure of PspF(1-275) R168A mutant | | Descriptor: | PSP OPERON TRANSCRIPTIONAL ACTIVATOR | | Authors: | Rappas, M, Schumacher, J, Beuron, F, Niwa, H, Bordes, P, Wigneshweraraj, S, Keetch, C.A, Robinson, C.V, Buck, M, Zhang, X. | | Deposit date: | 2005-02-08 | | Release date: | 2005-03-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Insights Into the Activity of Enhancer-Binding Proteins
Science, 307, 2005
|
|
2K2V
 
 | |
7XSN
 
 | | Native Tetrahymena ribozyme conformation | | Descriptor: | RNA (387-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSM
 
 | | Misfolded Tetrahymena ribozyme conformation 3 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (4.01 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSL
 
 | | Misfolded Tetrahymena ribozyme conformation 2 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.84 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7XSK
 
 | | Misfolded Tetrahymena ribozyme conformation 1 | | Descriptor: | RNA (388-MER) | | Authors: | Li, S, Palo, M, Pintilie, G, Zhang, X, Su, Z, Kappel, K, Chiu, W, Zhang, K, Das, R. | | Deposit date: | 2022-05-14 | | Release date: | 2022-08-03 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.53 Å) | | Cite: | Topological crossing in the misfolded Tetrahymena ribozyme resolved by cryo-EM.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YG8
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 5 undergoing the second-step self-splicing | | Descriptor: | MAGNESIUM ION, RNA (387-MER), RNA (5'-R(*CP*CP*CP*UP*C)-3'), ... | | Authors: | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YGA
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 2 undergoing the second-step self-splicing | | Descriptor: | MAGNESIUM ION, RNA (393-MER), RNA (5'-R(*CP*CP*CP*UP*CP*U)-3'), ... | | Authors: | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YG9
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 1 undergoing the second-step self-splicing | | Descriptor: | MAGNESIUM ION, RNA (391-MER), RNA (5'-R(*CP*CP*CP*UP*CP*U)-3'), ... | | Authors: | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YGD
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 6 undergoing the second-step self-splicing | | Descriptor: | MAGNESIUM ION, RNA (384-MER), RNA (5'-R(*CP*C)-3'), ... | | Authors: | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YGC
 
 | | Cryo-EM structure of Tetrahymena ribozyme conformation 4 undergoing the second-step self-splicing | | Descriptor: | MAGNESIUM ION, RNA (393-MER), RNA (5'-R(*CP*CP*CP*UP*CP*UP*UP*AP*AP*CP*C)-3'), ... | | Authors: | Li, S, Michael, Z.P, Zhang, X, Greg, P, Zhang, K. | | Deposit date: | 2022-07-11 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Snapshots of the second-step self-splicing of Tetrahymena ribozyme revealed by cryo-EM.
Nat Commun, 14, 2023
|
|
7YEZ
 
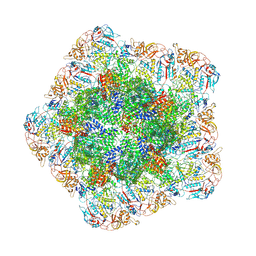 | | In situ structure of polymerase complex of mammalian reovirus in the reloaded state | | Descriptor: | Lambda-2 protein, Mu-2 protein, RNA helicase, ... | | Authors: | Bao, K.Y, Zhang, X.L, Li, D.Y, Zhu, P. | | Deposit date: | 2022-07-06 | | Release date: | 2023-03-29 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | In situ structures of polymerase complex of mammalian reovirus illuminate RdRp activation and transcription regulation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YF0
 
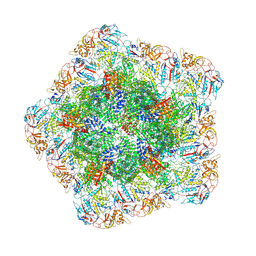 | | In situ structure of polymerase complex of mammalian reovirus in the core | | Descriptor: | Lambda-2 protein, Mu-2 protein, RNA helicase, ... | | Authors: | Bao, K.Y, Zhang, X.L, Li, D.Y, Zhu, P. | | Deposit date: | 2022-07-07 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | In situ structures of polymerase complex of mammalian reovirus illuminate RdRp activation and transcription regulation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YFE
 
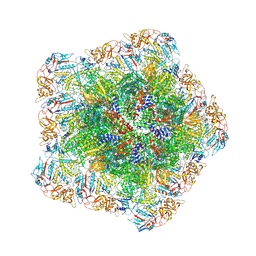 | | In situ structure of polymerase complex of mammalian reovirus in virion | | Descriptor: | Lambda-2 protein, Mu-2 protein, RNA (5'-R(P*AP*CP*GP*AP*UP*UP*AP*GP*C)-3'), ... | | Authors: | Bao, K.Y, Zhang, X.L, Li, D.Y, Zhu, P. | | Deposit date: | 2022-07-08 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | In situ structures of polymerase complex of mammalian reovirus illuminate RdRp activation and transcription regulation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YEV
 
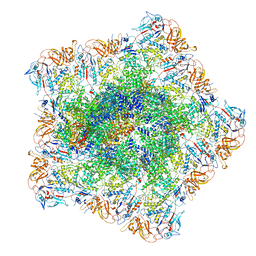 | | In situ structure of polymerase complex of mammalian reovirus in the pre-elongation state | | Descriptor: | Lambda-2 protein, Mu-2 protein, RNA helicase, ... | | Authors: | Bao, K.Y, Zhang, X.L, Li, D.Y, Zhu, P. | | Deposit date: | 2022-07-06 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | In situ structures of polymerase complex of mammalian reovirus illuminate RdRp activation and transcription regulation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7YED
 
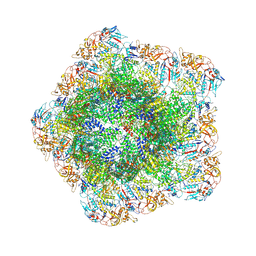 | | In situ structure of polymerase complex of mammalian reovirus in the elongation state | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Lambda-2 protein, MAGNESIUM ION, ... | | Authors: | Bao, K.Y, Zhang, X.L, Li, D.Y, Zhu, P. | | Deposit date: | 2022-07-05 | | Release date: | 2023-04-05 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | In situ structures of polymerase complex of mammalian reovirus illuminate RdRp activation and transcription regulation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
2GZY
 
 | |
7WL2
 
 | |
7WLE
 
 | |
7WL9
 
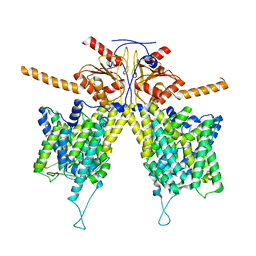 | | Mouse Pendrin in chloride and bicarbonate in asymmetric state | | Descriptor: | CHLORIDE ION, Pendrin | | Authors: | Liu, Q.Y, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2022-01-12 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Asymmetric pendrin homodimer reveals its molecular mechanism as anion exchanger.
Nat Commun, 14, 2023
|
|
7WK1
 
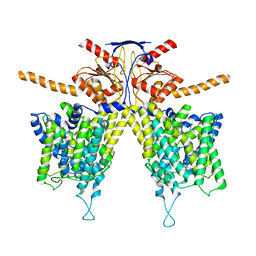 | | Mouse Pendrin bound chloride in inward state | | Descriptor: | CHLORIDE ION, Pendrin | | Authors: | Liu, Q.Y, Zhang, X, Sun, L, Chen, Z.G. | | Deposit date: | 2022-01-08 | | Release date: | 2023-04-12 | | Last modified: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Asymmetric pendrin homodimer reveals its molecular mechanism as anion exchanger.
Nat Commun, 14, 2023
|
|
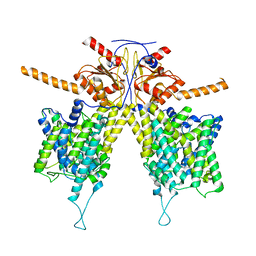
7WLA
 
 | |
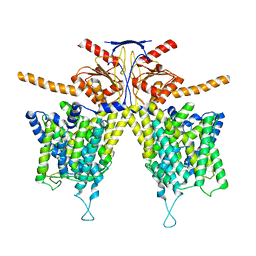
7WL7
 
 | |
2GZZ
 
 | |
