8H83
 
 | | Crystal structure of a IsPETase variant V22 from Ideonella sakaiensis | | Descriptor: | Poly(ethylene terephthalate) hydrolase | | Authors: | Wei, H.L, Gao, S.F, Li, Q, Han, X, Gao, J, Liu, W.D. | | Deposit date: | 2022-10-21 | | Release date: | 2024-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Crystal structure of a IsPETase variant V22 from Ideonella sakaiensis
to be published
|
|
7CUV
 
 | |
8H8Y
 
 | | Crystal structure of AbHheG from Acidimicrobiia bacterium | | Descriptor: | GLYCEROL, alpha/beta hydrolase | | Authors: | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | Deposit date: | 2022-10-24 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
3SW9
 
 | | GLP (G9a-like protein) SET domain in complex with Dnmt3aK44me0 peptide | | Descriptor: | DNA (cytosine-5)-methyltransferase 3A, Histone-lysine N-methyltransferase EHMT1, SINEFUNGIN, ... | | Authors: | Chang, Y, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2011-07-13 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | MPP8 mediates the interactions between DNA methyltransferase Dnmt3a and H3K9 methyltransferase GLP/G9a.
Nat Commun, 2, 2011
|
|
3SVM
 
 | | Human MPP8 - human DNMT3AK47me2 peptide | | Descriptor: | DNA (cytosine-5)-methyltransferase 3A, M-phase phosphoprotein 8 | | Authors: | Chang, Y, Horton, J.R, Zhang, X, Cheng, X. | | Deposit date: | 2011-07-12 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | MPP8 mediates the interactions between DNA methyltransferase Dnmt3a and H3K9 methyltransferase GLP/G9a.
Nat Commun, 2, 2011
|
|
6H6H
 
 | |
8IVP
 
 | | Crystal structure of MV in complex with LLP and FRU from Mycobacterium vanbaalenii | | Descriptor: | Branched chain amino acid: 2-keto-4-methylthiobutyrate aminotransferase, D-fructose | | Authors: | Li, Q, Zhu, Y.M, Gao, J, Wei, H.L, Han, X, Liu, W.D, Sun, Y.X. | | Deposit date: | 2023-03-28 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Crystal structure of MV in complex with LLP and FRU from Mycobacterium vanbaalenii
To Be Published
|
|
6H6D
 
 | |
8II9
 
 | | crystal structure of Hyp mutant from Hypoxylon sp. E7406B | | Descriptor: | Terpene synthase | | Authors: | Gao, J, Liu, W.D, Li, Q, Han, X, Wei, H.L, Dai, Z.J, Su, L.Q. | | Deposit date: | 2023-02-24 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | crystal structure of Hyp
to be published
|
|
8IJT
 
 | | crystal structure of Hyp N135A mutant from Hypoxylon sp. E7406B | | Descriptor: | Terpene synthase | | Authors: | Gao, J, Su, L.Q, Li, Q, Han, X, Wei, H.L, Dai, Z.J, Liu, W.D. | | Deposit date: | 2023-02-28 | | Release date: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | crystal structure of Hyp N135A mutant from Hypoxylon sp. E7406B
to be published
|
|
8HQP
 
 | | Crystal structure of AbHheG mutant from Acidimicrobiia bacterium | | Descriptor: | AbHheG_m | | Authors: | Zhou, C.H, Chen, X, Han, X, Liu, W.D, Wu, Q.Q, Zhu, D.M, Ma, Y.H. | | Deposit date: | 2022-12-13 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Flipping the Substrate Creates a Highly Selective Halohydrin Dehalogenase for the Synthesis of Chiral 4-Aryl-2-oxazolidinones from Readily Available Epoxides
Acs Catalysis, 13, 2023
|
|
5XNN
 
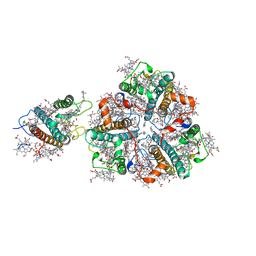 | | Structure of M-LHCII and CP24 complexes in the stacked C2S2M2-type PSII-LHCII supercomplex from Pisum sativum | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Su, X.D, Ma, J, Wei, X.P, Cao, P, Zhu, D.J, Chang, W.R, Liu, Z.F, Zhang, X.Z, Li, M. | | Deposit date: | 2017-05-23 | | Release date: | 2017-09-20 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex
Science, 357, 2017
|
|
5YCO
 
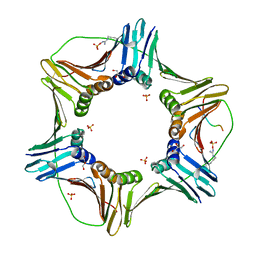 | | Complex structure of PCNA with UHRF2 | | Descriptor: | E3 ubiquitin-protein ligase UHRF2, GLYCEROL, Proliferating cell nuclear antigen, ... | | Authors: | Wu, M, Chen, W, Hang, T, Wang, C, Zhang, X, Zang, J. | | Deposit date: | 2017-09-07 | | Release date: | 2017-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.199 Å) | | Cite: | Structure insights into the molecular mechanism of the interaction between UHRF2 and PCNA.
Biochem. Biophys. Res. Commun., 494, 2017
|
|
6U6V
 
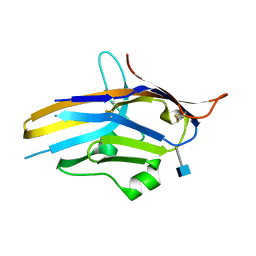 | | Crystal structure of human PD-1H / VISTA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, V-type immunoglobulin domain-containing suppressor of T-cell activation | | Authors: | Slater, B.T, Han, X, Chen, L, Xiong, Y. | | Deposit date: | 2019-08-30 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insight into T cell coinhibition by PD-1H (VISTA).
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
3WUG
 
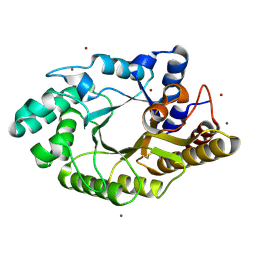 | | The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) with xylobiose from Streptomyces sp. 9 | | Descriptor: | Endo-1,4-beta-xylanase A, ZINC ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y.Y, Guo, R.T. | | Deposit date: | 2014-04-23 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
3WUE
 
 | | The wild type crystal structure of b-1,4-Xylanase (XynAS9) with xylobiose from Streptomyces sp. 9 | | Descriptor: | Endo-1,4-beta-xylanase A, ZINC ION, beta-D-xylopyranose-(1-4)-beta-D-xylopyranose | | Authors: | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y, Guo, R.T. | | Deposit date: | 2014-04-23 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
3WUB
 
 | | The wild type crystal structure of b-1,4-Xylanase (XynAS9) from Streptomyces sp. 9 | | Descriptor: | Endo-1,4-beta-xylanase A, ZINC ION | | Authors: | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y.Y, Guo, R.T. | | Deposit date: | 2014-04-23 | | Release date: | 2014-10-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
3WUF
 
 | | The mutant crystal structure of b-1,4-Xylanase (XynAS9_V43P/G44E) from Streptomyces sp. 9 | | Descriptor: | Endo-1,4-beta-xylanase A, ZINC ION | | Authors: | Chen, C.C, Han, X, Lv, P, Ko, T.P, Peng, W, Huang, C.H, Zheng, Y, Gao, J, Yang, Y.Y, Guo, R.T. | | Deposit date: | 2014-04-23 | | Release date: | 2014-10-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural perspectives of an engineered beta-1,4-xylanase with enhanced thermostability.
J.Biotechnol., 189C, 2014
|
|
1XOO
 
 | | NMR structure of G1S mutant of influenza hemagglutinin fusion peptide in DPC micelles at pH 5 | | Descriptor: | Hemagglutinin | | Authors: | Li, Y, Han, X, Lai, A.L, Bushweller, J.H, Cafiso, D.S, Tamm, L.K. | | Deposit date: | 2004-10-06 | | Release date: | 2005-09-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Membrane structures of the hemifusion-inducing fusion peptide mutant G1S and the fusion-blocking mutant G1V of influenza virus hemagglutinin suggest a mechanism for pore opening in membrane fusion.
J.Virol., 79, 2005
|
|
5M00
 
 | | Crystal structure of murine P14 TCR complex with H-2Db and Y4A, modified gp33 peptide from LCMV | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Achour, A, Sandalova, T, Sun, R, Han, X. | | Deposit date: | 2016-10-03 | | Release date: | 2017-12-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Thernary complexes of TCR P14 give insights into the mechanisms behind reestablishment of CTL responses against a viral escape mutant
To Be Published
|
|
5M01
 
 | | Crystal structure of murine P14 TCR/ H-2Db complex with PA, modified gp33 peptide from LCMV | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Achour, A, Sandalova, T, Sun, R, Han, X. | | Deposit date: | 2016-10-03 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Thernary complexes of TCR P14 give insights into the mechanisms behind reestablishment of CTL responses against a viral escape mutant
To Be Published
|
|
1XOP
 
 | | NMR structure of G1V mutant of influenza hemagglutinin fusion peptide in DPC micelles at pH 5 | | Descriptor: | Hemagglutinin | | Authors: | Li, Y, Han, X, Lai, A.L, Bushweller, J.H, Cafiso, D.S, Tamm, L.K. | | Deposit date: | 2004-10-06 | | Release date: | 2005-09-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Membrane structures of the hemifusion-inducing fusion peptide mutant G1S and the fusion-blocking mutant G1V of influenza virus hemagglutinin suggest a mechanism for pore opening in membrane fusion.
J.Virol., 79, 2005
|
|
7VPA
 
 | | Crystal structure of Ple629 from marine microbial consortium | | Descriptor: | hydrolase Ple629 | | Authors: | Wu, P, Zhao, Y.P, Li, Z.S, Ingrid, M.C, Lara, P, Gao, J, Han, X, Li, Q, Basak, O, Liu, W.D, Wei, R. | | Deposit date: | 2021-10-15 | | Release date: | 2022-08-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular and Biochemical Differences of the Tandem and Cold-Adapted PET Hydrolases Ple628 and Ple629, Isolated From a Marine Microbial Consortium.
Front Bioeng Biotechnol, 10, 2022
|
|
7VMD
 
 | | Crystal structure of a hydrolases Ple628 from marine microbial consortium | | Descriptor: | CALCIUM ION, hydrolase Ple628 | | Authors: | Wu, P, Zhao, Y.P, Li, Z.S, Ingrid, M.C, Lara, P, Gao, J, Han, X, Li, Q, Basak, O, Liu, W.D, Wei, R. | | Deposit date: | 2021-10-08 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular and Biochemical Differences of the Tandem and Cold-Adapted PET Hydrolases Ple628 and Ple629, Isolated From a Marine Microbial Consortium.
Front Bioeng Biotechnol, 10, 2022
|
|
7W66
 
 | | Crystal structure of a PSH1 mutant in complex with ligand | | Descriptor: | PSH1, bis(2-hydroxyethyl) benzene-1,4-dicarboxylate | | Authors: | Gao, J, Lara, P, Li, Z.S, Han, X, Wei, R, Liu, W.D. | | Deposit date: | 2021-12-01 | | Release date: | 2022-09-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Multiple Substrate Binding Mode-Guided Engineering of a Thermophilic PET Hydrolase.
Acs Catalysis, 12, 2022
|
|
