6VBG
 
 | | Lactose permease complex with thiodigalactoside and nanobody 9043 | | Descriptor: | Galactoside permease, beta-D-galactopyranose-(1-1)-1-thio-beta-D-galactopyranose, nanobody 9043, ... | | Authors: | Kumar, H, Stroud, R.M, Kaback, H.R, Finer-Moore, J, Smirnova, I, Kasho, V, Pardon, E, Steyart, J. | | Deposit date: | 2019-12-18 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Diversity in kinetics correlated with structure in nano body-stabilized LacY.
Plos One, 15, 2020
|
|
2Y5Y
 
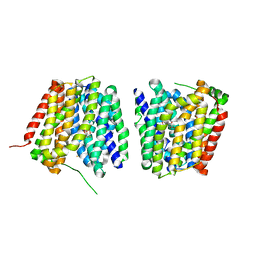 | | Crystal structure of LacY in complex with an affinity inactivator | | Descriptor: | 2-sulfanylethyl beta-D-galactopyranoside, BARIUM ION, LACTOSE PERMEASE | | Authors: | Chaptal, V, Kwon, S, Sawaya, M.R, Guan, L, Kaback, H.R, Abramson, J. | | Deposit date: | 2011-01-19 | | Release date: | 2011-06-15 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Crystal Structure of Lactose Permease in Complex with an Affinity Inactivator Yields Unique Insight Into Sugar Recognition.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
1CGO
 
 | | CYTOCHROME C' | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Dobbs, A.J, Faber, H.R, Anderson, B.F, Baker, E.N. | | Deposit date: | 1995-05-01 | | Release date: | 1995-07-31 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Three-dimensional structure of cytochrome c' from two Alcaligenes species and the implications for four-helix bundle structures.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
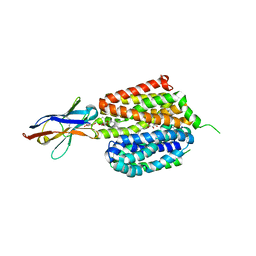
1BU6
 
 | | CRYSTAL STRUCTURES OF ESCHERICHIA COLI GLYCEROL KINASE AND THE MUTANT A65T IN AN INACTIVE TETRAMER: CONFORMATIONAL CHANGES AND IMPLICATIONS FOR ALLOSTERIC REGULATION | | Descriptor: | GLYCEROL, PROTEIN (GLYCEROL KINASE), SULFATE ION | | Authors: | Feese, M.D, Faber, H.R, Bystrom, C.E, Pettigrew, D.W, Remington, S.J. | | Deposit date: | 1998-08-30 | | Release date: | 1998-09-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Glycerol kinase from Escherichia coli and an Ala65-->Thr mutant: the crystal structures reveal conformational changes with implications for allosteric regulation.
Structure, 6, 1998
|
|
2OFN
 
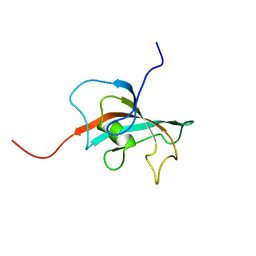 | | Solution structure of FK506-binding domain (FKBD)of FKBP35 from Plasmodium falciparum | | Descriptor: | 70 kDa peptidylprolyl isomerase, putative | | Authors: | Kang, C.B, Ye, H, Yoon, H.R, Yoon, H.S. | | Deposit date: | 2007-01-04 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution structure of FK506 binding domain (FKBD) of Plasmodium falciparum FK506 binding protein 35 (PfFKBP35).
Proteins, 70, 2007
|
|
6TFR
 
 | | Linalool Dehydratase Isomerase C180A mutant | | Descriptor: | 1,2-ETHANEDIOL, Linalool dehydratase-isomerase protein LDI | | Authors: | Cuetos, A, Zukic, E, Danesh-Azari, H.R, Grogan, G. | | Deposit date: | 2019-11-14 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Mutational Analysis of Linalool Dehydratase Isomerase Suggests That Alcohol and Alkene Transformations Are Catalyzed Using Noncovalent Mechanisms
Acs Catalysis, 2020
|
|
6TFT
 
 | |
1DSN
 
 | | D60S N-TERMINAL LOBE HUMAN LACTOFERRIN | | Descriptor: | CARBONATE ION, FE (III) ION, LACTOFERRIN | | Authors: | Faber, H.R, Norris, G.E, Baker, E.N. | | Deposit date: | 1995-12-13 | | Release date: | 1996-03-08 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Altered domain closure and iron binding in transferrins: the crystal structure of the Asp60Ser mutant of the amino-terminal half-molecule of human lactoferrin.
J.Mol.Biol., 256, 1996
|
|
2PHE
 
 | | Model for VP16 binding to PC4 | | Descriptor: | Alpha trans-inducing protein, TRANSCRIPTIONAL COACTIVATOR PC4 | | Authors: | Jonker, H.R.A, Wechselberger, R.W, Boelens, R, Folkers, G.E, Kaptein, R. | | Deposit date: | 2007-04-11 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Properties of the Promiscuous VP16 Activation Domain
Biochemistry, 44, 2005
|
|
1CGN
 
 | | CYTOCHROME C' | | Descriptor: | CYTOCHROME C, HEME C | | Authors: | Dobbs, A.J, Faber, H.R, Anderson, B.F, Baker, E.N. | | Deposit date: | 1995-05-01 | | Release date: | 1995-07-31 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Three-dimensional structure of cytochrome c' from two Alcaligenes species and the implications for four-helix bundle structures.
Acta Crystallogr.,Sect.D, 52, 1996
|
|
2YMM
 
 | | Sulfate bound L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | L-HALOACID DEHALOGENASE, SULFATE ION | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-09 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
2PHG
 
 | | Model for VP16 binding to TFIIB | | Descriptor: | Alpha trans-inducing protein, Transcription initiation factor IIB | | Authors: | Jonker, H.R.A, Wechselberger, R.W, Boelens, R, Folkers, G.E, Kaptein, R. | | Deposit date: | 2007-04-11 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Properties of the Promiscuous VP16 Activation Domain
Biochemistry, 44, 2005
|
|
2YML
 
 | | Native L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | L-HALOACID DEHALOGENASE | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-09 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
2YMP
 
 | | Chloroacetic acid complex bound L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | L-HALOACID DEHALOGENASE | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
2YMQ
 
 | | Chloropropionic acid complex bound L-haloacid dehalogenase from a Rhodobacteraceae family bacterium | | Descriptor: | L-HALOACID DEHALOGENASE | | Authors: | Novak, H.R, Sayer, C, Isupov, M.N, Paszkiewicz, K, Gotz, D, Spragg, A.M, Littlechild, J.A. | | Deposit date: | 2012-10-10 | | Release date: | 2013-05-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Marine Rhodobacteraceae L-Haloacid Dehalogenase Contains a Novel His/Glu Dyad that Could Activate the Catalytic Water.
FEBS J., 280, 2013
|
|
2RGF
 
 | | RBD OF RAL GUANOSINE-NUCLEOTIDE EXCHANGE FACTOR (PROTEIN), NMR, 10 STRUCTURES | | Descriptor: | RALGEF-RBD | | Authors: | Geyer, M, Herrmann, C, Wittinghofer, A, Kalbitzer, H.R. | | Deposit date: | 1997-02-13 | | Release date: | 1998-03-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Ras-binding domain of RalGEF and implications for Ras binding and signalling.
Nat.Struct.Biol., 4, 1997
|
|
2W0G
 
 | |
2OBX
 
 | | Lumazine synthase RibH2 from Mesorhizobium loti (Gene mll7281, Swiss-Prot entry Q986N2) complexed with inhibitor 5-Nitro-6-(D-Ribitylamino)-2,4(1H,3H) Pyrimidinedione | | Descriptor: | 5-NITRO-6-RIBITYL-AMINO-2,4(1H,3H)-PYRIMIDINEDIONE, 6,7-dimethyl-8-ribityllumazine synthase 1, PHOSPHATE ION | | Authors: | Klinke, S, Zylberman, V, Bonomi, H.R, Haase, I, Guimaraes, B.G, Braden, B.C, Bacher, A, Fischer, M, Goldbaum, F.A. | | Deposit date: | 2006-12-20 | | Release date: | 2007-08-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Structural and kinetic properties of lumazine synthase isoenzymes in the order rhizobiales
J.Mol.Biol., 373, 2007
|
|
2UZ2
 
 | | Crystal structure of Xenavidin | | Descriptor: | ACETATE ION, BIOTIN, XENAVIDIN | | Authors: | Helppolainen, S.H, Maatta, J.A.E, Airenne, T.T, Johnson, M.S, Kulomaa, M.S, Nordlund, H.R. | | Deposit date: | 2007-04-24 | | Release date: | 2008-06-03 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Characteristics of Xenavidin, the First Frog Avidin from Xenopus Tropicalis.
Bmc Struct.Biol., 9, 2009
|
|
1E0A
 
 | | Cdc42 complexed with the GTPase binding domain of p21 activated kinase | | Descriptor: | Cell division control protein 42 homolog, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Morreale, A, Venkatesan, M, Mott, H.R, Owen, D, Nietlispach, D, Lowe, P.N, Laue, E.D. | | Deposit date: | 2000-03-16 | | Release date: | 2000-04-18 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Cdc42 Bound to the Gtpase Binding Domian of Pak
Nat.Struct.Biol., 7, 2000
|
|
2ZLE
 
 | | Cryo-EM structure of DegP12/OMP | | Descriptor: | Outer membrane protein C, Protease do | | Authors: | Schaefer, E, Saibil, H.R. | | Deposit date: | 2008-04-09 | | Release date: | 2008-06-03 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (28 Å) | | Cite: | Structural basis for the regulated protease and chaperone function of DegP
Nature, 453, 2008
|
|
344D
 
 | | DETERMINATION BY MAD-DM OF THE STRUCTURE OF THE DNA DUPLEX D(ACGTACG(5-BRU))2 AT 1.46A AND 100K | | Descriptor: | DNA (5'-D(*AP*CP*GP*TP*AP*CP*GP*(BRU))-3') | | Authors: | Todd, A.R, Adams, A, Powell, H.R, Cardin, C.J. | | Deposit date: | 1997-08-04 | | Release date: | 1997-09-26 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Determination by MAD-DM of the structure of the DNA duplex d[ACGTACG(5-BrU)]2 at 1.46 A and 100 K.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1B4I
 
 | | Control of K+ Channel Gating by protein phosphorylation: structural switches of the inactivation gate, NMR, 22 structures | | Descriptor: | POTASSIUM CHANNEL | | Authors: | Antz, C, Bauer, T, Kalbacher, H, Frank, R, Covarrubias, M, Kalbitzer, H.R, Ruppersberg, J.P, Baukrowitz, T, Fakler, B. | | Deposit date: | 1998-12-22 | | Release date: | 1999-04-27 | | Last modified: | 2022-03-23 | | Method: | SOLUTION NMR | | Cite: | Control of K+ channel gating by protein phosphorylation: structural switches of the inactivation gate.
Nat.Struct.Biol., 6, 1999
|
|
2VID
 
 | | Serine protease SplB from Staphylococcus aureus at 1.8A resolution | | Descriptor: | SERINE PROTEASE SPLB | | Authors: | Dubin, G, Stec-Niemczyk, J, Kisielewska, M, Pustelny, K, Popowicz, G.M, Bista, M, Kantyka, T, Boulware, K.T, Stennicke, H.R, Czarna, A, Phopaisarn, M, Daugherty, P.S, Thogersen, I.B, Enghild, J.J, Thornberry, N, Dubin, A, Potempa, J. | | Deposit date: | 2007-11-30 | | Release date: | 2008-05-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Enzymatic Activity of the Staphylococcus Aureus Splb Serine Protease is Induced by Substrates Containing the Sequence Trp-Glu-Leu-Gln.
J.Mol.Biol., 379, 2008
|
|
2UYW
 
 | | Crystal structure of Xenavidin | | Descriptor: | BIOTIN, FORMIC ACID, XENAVIDIN | | Authors: | Helppolainen, S.H, Maatta, J.A.E, Airenne, T.T, Johnson, M.S, Kulomaa, M.S, Nordlund, H.R. | | Deposit date: | 2007-04-20 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Characteristics of Xenavidin, the First Frog Avidin from Xenopus Tropicalis.
Bmc Struct.Biol., 9, 2009
|
|
