4V3M
 
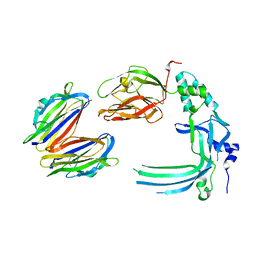 | | Membrane bound pleurotolysin prepore (TMH2 helix lock) trapped with engineered disulphide cross-link | | Descriptor: | PLEUROTOLYSIN A, PLEUROTOLYSIN B | | Authors: | Lukoyanova, N, Kondos, S.C, Farabella, I, Law, R.H.P, Reboul, C.F, Caradoc-Davies, T.T, Spicer, B.A, Kleifeld, O, Perugini, M, Ekkel, S, Hatfaludi, T, Oliver, K, Hotze, E.M, Tweten, R.K, Whisstock, J.C, Topf, M, Dunstone, M.A, Saibil, H.R. | | Deposit date: | 2014-10-20 | | Release date: | 2015-02-18 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (17 Å) | | Cite: | Conformational Changes During Pore Formation by the Perforin-Related Protein Pleurotolysin.
Plos Biol., 13, 2015
|
|
6ZR2
 
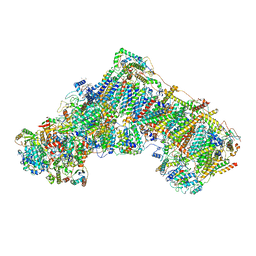 | | Cryo-EM structure of respiratory complex I in the active state from Mus musculus at 3.1 A | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOETHANOLAMINE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Bridges, H.R, Blaza, J.N, Agip, A.N.A, Hirst, J. | | Deposit date: | 2020-07-10 | | Release date: | 2020-10-21 | | Last modified: | 2020-10-28 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of inhibitor-bound mammalian complex I.
Nat Commun, 11, 2020
|
|
8IDZ
 
 | |
8I1V
 
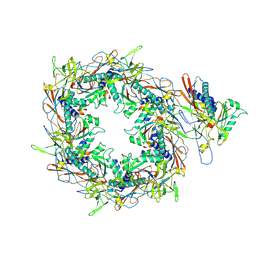 | | The asymmetric unit of P22 procapsid | | Descriptor: | Major capsid protein, Scaffolding protein | | Authors: | Xiao, H, Liu, H.R, Cheng, L.P. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-08 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Assembly and Capsid Expansion Mechanism of Bacteriophage P22 Revealed by High-Resolution Cryo-EM Structures.
Viruses, 15, 2023
|
|
8I1T
 
 | | The asymmetric unit of P22 empty capsid | | Descriptor: | Major capsid protein | | Authors: | Xiao, H, Liu, H.R, Cheng, L.P. | | Deposit date: | 2023-01-13 | | Release date: | 2023-03-08 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Assembly and Capsid Expansion Mechanism of Bacteriophage P22 Revealed by High-Resolution Cryo-EM Structures.
Viruses, 15, 2023
|
|
6IDM
 
 | | Crystal structure of Peptidoglycan recognition protein (PGRP-S) with Tartaric acid at 3.20 A resolution | | Descriptor: | L(+)-TARTARIC ACID, Peptidoglycan recognition protein 1 | | Authors: | Bairagya, H.R, Shokeen, A, Sharma, P, Singh, P.K, Sharma, S, Singh, T.P. | | Deposit date: | 2018-09-10 | | Release date: | 2018-09-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of Peptidoglycan recognition protein (PGRP-S) with Tartaric acid at 3.20 A resolution
To Be Published
|
|
4NBU
 
 | | Crystal structure of FabG from Bacillus sp | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 3-oxoacyl-(Acyl-carrier-protein) reductase, ACETOACETYL-COENZYME A | | Authors: | Pereira, J.H, Mcandrew, R.P, Javidpour, P, Beller, H.R, Adams, P.D. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Biochemical and Structural Studies of NADH-Dependent FabG Used To Increase the Bacterial Production of Fatty Acids under Anaerobic Conditions.
Appl.Environ.Microbiol., 80, 2014
|
|
4NBT
 
 | | Crystal structure of FabG from Acholeplasma laidlawii | | Descriptor: | 3-oxoacyl-[acyl-carrier-protein] reductase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Pereira, J.H, Mcandrew, R.P, Javidpour, P, Beller, H.R, Adams, P.D. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Biochemical and Structural Studies of NADH-Dependent FabG Used To Increase the Bacterial Production of Fatty Acids under Anaerobic Conditions.
Appl.Environ.Microbiol., 80, 2014
|
|
4NBV
 
 | | Crystal structure of FabG from Cupriavidus taiwanensis | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-oxoacyl-[acyl-carrier-protein] reductase putative short-chain dehydrogenases/reductases (SDR) family protein | | Authors: | Pereira, J.H, Mcandrew, R.P, Javidpour, P, Beller, H.R, Adams, P.D. | | Deposit date: | 2013-10-23 | | Release date: | 2013-12-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.645 Å) | | Cite: | Biochemical and Structural Studies of NADH-Dependent FabG Used To Increase the Bacterial Production of Fatty Acids under Anaerobic Conditions.
Appl.Environ.Microbiol., 80, 2014
|
|
6JJQ
 
 | | Crystal structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii at 0.99 A resolution. | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Peptidyl-tRNA hydrolase, ... | | Authors: | Viswanathan, V, Bairagya, H.R, Sharma, P, Sharma, S, Singh, T.P. | | Deposit date: | 2019-02-26 | | Release date: | 2019-03-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (0.99 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase from Acinetobacter baumannii at 0.99 A resolution.
To Be Published
|
|
4ZYR
 
 | | Crystal structure of E. coli Lactose permease G46W/G262W bound to p-nitrophenyl alpha-D-galactopyranoside (alpha-NPG) | | Descriptor: | 4-nitrophenyl alpha-D-galactopyranoside, Lactose permease, nonyl beta-D-glucopyranoside | | Authors: | Kumar, H, Finer-Moore, J.S, Kaback, H.R, Stroud, R.M. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.312 Å) | | Cite: | Structure of LacY with an alpha-substituted galactoside: Connecting the binding site to the protonation site.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4ZTT
 
 | | Crystal structures of ferritin mutants reveal diferric-peroxo intermediates | | Descriptor: | Bacterial non-heme ferritin, FE (II) ION, FE (III) ION, ... | | Authors: | Kim, S, Park, Y.H, Jung, S.W, Seok, J.H, Chung, Y.B, Lee, D.B, Gowda, G, Lee, J.H, Han, H.R, Cho, A.E, Lee, C, Chung, M.S, Kim, K.H. | | Deposit date: | 2015-05-15 | | Release date: | 2016-06-15 | | Last modified: | 2020-02-19 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis of Novel Iron-Uptake Route and Reaction Intermediates in Ferritins from Gram-Negative Bacteria.
J. Mol. Biol., 428, 2016
|
|
6JOG
 
 | | Crystal structure of the complex of phospho pantetheine adenylyl transferase from Acinetobacter baumannii with two ascorbic acid (Vitamin-C) molecules. | | Descriptor: | ASCORBIC ACID, Phosphopantetheine adenylyltransferase, SULFATE ION | | Authors: | Viswanathan, V, Gupta, A, Bairagya, H.R, Sharma, S, Singh, T.P. | | Deposit date: | 2019-03-20 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the complex of phospho pantetheine adenylyl transferase from Acinetobacter baumannii with two ascorbic acid (Vitamin-C) molecules.
To Be Published
|
|
5AKP
 
 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP from Xanthomonas campestris bound to BV chromophore | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BILIVERDINE IX ALPHA, CHLORIDE ION, ... | | Authors: | Otero, L.H, Klinke, S, Goldbaum, F.A, Bonomi, H.R. | | Deposit date: | 2015-03-04 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure of the Full-Length Bacteriophytochrome from the Plant Pathogen Xanthomonas Campestris Provides Clues to its Long-Range Signaling Mechanism.
J.Mol.Biol., 428, 2016
|
|
8ZKM
 
 | |
9IN6
 
 | | Capsid of Vibrio cholerae phage mature VP1 | | Descriptor: | SHP of VP1, major capsid of VP1 | | Authors: | Liu, H.R, Pang, H. | | Deposit date: | 2024-07-05 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Three-dimensional structures of Vibrio cholerae typing podophage VP1 in two states
to be published
|
|
8ZKK
 
 | |
6LZM
 
 | | COMPARISON OF THE CRYSTAL STRUCTURE OF BACTERIOPHAGE T4 LYSOZYME AT LOW, MEDIUM, AND HIGH IONIC STRENGTHS | | Descriptor: | BETA-MERCAPTOETHANOL, CHLORIDE ION, T4 LYSOZYME | | Authors: | Bell, J.A, Wilson, K, Zhang, X.-J, Faber, H.R, Nicholson, H, Matthews, B.W. | | Deposit date: | 1991-01-25 | | Release date: | 1992-07-15 | | Last modified: | 2021-06-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Comparison of the crystal structure of bacteriophage T4 lysozyme at low, medium, and high ionic strengths.
Proteins, 10, 1991
|
|
1TXE
 
 | | Solution structure of the active-centre mutant Ile14Ala of the histidine-containing phosphocarrier protein (HPr) from Staphylococcus carnosus | | Descriptor: | Phosphocarrier protein HPr | | Authors: | Moeglich, A, Koch, B, Hengstenberg, W, Brunner, E, Kalbitzer, H.R, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-07-04 | | Release date: | 2005-03-08 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the active-centre mutant I14A of the histidine-containing phosphocarrier protein from Staphylococcus carnosus
Eur.J.Biochem., 271, 2004
|
|
1UB1
 
 | | Solution structure of the matrix attachment region-binding domain of chicken MeCP2 | | Descriptor: | attachment region binding protein | | Authors: | Heitmann, B, Maurer, T, Weitzel, J.M, Stratling, W.H, Kalbitzer, H.R, Brunner, E, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2003-03-27 | | Release date: | 2003-08-05 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the matrix attachment region-binding domain of chicken MeCP2
EUR.J.BIOCHEM., 270, 2003
|
|
1URF
 
 | | HR1b domain from PRK1 | | Descriptor: | PROTEIN KINASE C-LIKE 1 | | Authors: | Owen, D, Lowe, P.N, Nietlispach, D, Brosnan, C.E, Chirgadze, D.Y, Parker, P.J, Blundell, T.L, Mott, H.R. | | Deposit date: | 2003-10-29 | | Release date: | 2003-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Dissection of the Interaction between the Small G Proteins Rac1 and Rhoa and Protein Kinase C-Related Kinase 1 (Prk1)
J.Biol.Chem., 278, 2003
|
|
1UQV
 
 | | SAM domain from Ste50p | | Descriptor: | STE50 PROTEIN | | Authors: | Grimshaw, S.J, Mott, H.R, Stott, K.M, Nielsen, P.R, Evetts, K.A, Hopkins, L.J, Nietlispach, D, Owen, D. | | Deposit date: | 2003-10-20 | | Release date: | 2003-10-30 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure of the Sterile {Alpha} Motif (Sam) Domain of the Saccharomyces Cerevisiae Mitogen-Activated Protein Kinase Pathway-Modulating Protein Ste50 and Analysis of its Interaction with the Ste11 Sam
J.Biol.Chem., 279, 2004
|
|
6VBG
 
 | | Lactose permease complex with thiodigalactoside and nanobody 9043 | | Descriptor: | Galactoside permease, beta-D-galactopyranose-(1-1)-1-thio-beta-D-galactopyranose, nanobody 9043, ... | | Authors: | Kumar, H, Stroud, R.M, Kaback, H.R, Finer-Moore, J, Smirnova, I, Kasho, V, Pardon, E, Steyart, J. | | Deposit date: | 2019-12-18 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Diversity in kinetics correlated with structure in nano body-stabilized LacY.
Plos One, 15, 2020
|
|
5UYR
 
 | | Crystal structure of the dark-adapted full-length bacteriophytochrome XccBphP mutant D199A from Xanthomonas campestris | | Descriptor: | BILIVERDINE IX ALPHA, Bacteriophytochrome | | Authors: | Otero, L.H, Klinke, S, Goldbaum, F.A, Bonomi, H.R. | | Deposit date: | 2017-02-24 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.45 Å) | | Cite: | Pr-favoured variants of the bacteriophytochrome from the plant pathogen Xanthomonas campestris hint on light regulation of virulence-associated mechanisms.
Febs J., 288, 2021
|
|
6F27
 
 | | NMR solution structure of non-bound [des-Arg10]-kallidin (DAKD) | | Descriptor: | DAKD | | Authors: | Richter, C, Jonker, H.R.A, Schwalbe, H, Joedicke, L, Mao, J, Kuenze, G, Reinhart, C, Kalavacherla, T, Meiler, J, Preu, J, Michel, H, Glaubitz, C. | | Deposit date: | 2017-11-23 | | Release date: | 2018-01-10 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors.
Nat. Chem. Biol., 14, 2018
|
|
