4B0T
 
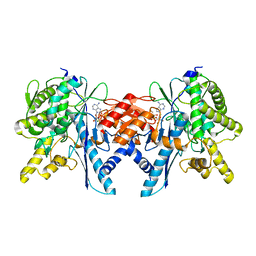 | | Structure of the Pup Ligase PafA of the Prokaryotic Ubiquitin-like Modification Pathway in Complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PUP--PROTEIN LIGASE | | Authors: | Ozcelik, D, Barandun, J, Schmitz, N, Sutter, M, Guth, E, Damberger, F.F, Allain, F.H.-T, Ban, N, Weber-Ban, E. | | Deposit date: | 2012-07-04 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.159 Å) | | Cite: | Structures of Pup Ligase Pafa and Depupylase Dop from the Prokaryotic Ubiquitin-Like Modification Pathway.
Nat.Commun., 3, 2012
|
|
4B0S
 
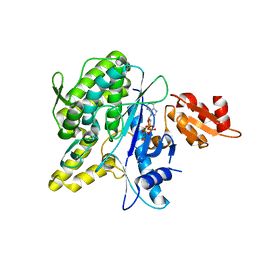 | | Structure of the Deamidase-Depupylase Dop of the Prokaryotic Ubiquitin-like Modification Pathway in Complex with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DEAMIDASE-DEPUPYLASE DOP, MAGNESIUM ION | | Authors: | Ozcelik, D, Barandun, J, Schmitz, N, Sutter, M, Guth, E, Damberger, F.F, Allain, F.H.-T, Ban, N, Weber-Ban, E. | | Deposit date: | 2012-07-04 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structures of Pup Ligase Pafa and Depupylase Dop from the Prokaryotic Ubiquitin-Like Modification Pathway.
Nat.Commun., 3, 2012
|
|
4B0R
 
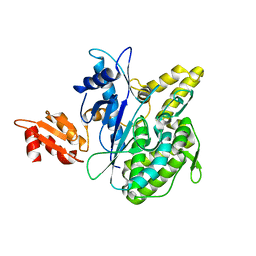 | | Structure of the Deamidase-Depupylase Dop of the Prokaryotic Ubiquitin-like Modification Pathway | | Descriptor: | DEAMIDASE-DEPUPYLASE DOP | | Authors: | Ozcelik, D, Barandun, J, Schmitz, N, Sutter, M, Guth, E, Damberger, F.F, Allain, F.H.-T, Ban, N, Weber-Ban, E. | | Deposit date: | 2012-07-04 | | Release date: | 2012-09-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structures of Pup ligase PafA and depupylase Dop from the prokaryotic ubiquitin-like modification pathway.
Nat Commun, 3, 2012
|
|
1LNL
 
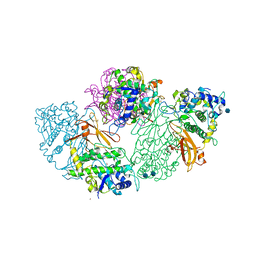 | | Structure of deoxygenated hemocyanin from Rapana thomasiana | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COPPER (II) ION, hemocyanin | | Authors: | Perbandt, M, Guthoehrlein, E.W, Rypniewski, W, Idakieva, K, Stoeva, S, Voelter, W, Genov, N, Betzel, C. | | Deposit date: | 2002-05-03 | | Release date: | 2003-06-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | The structure of a functional unit from the wall of a gastropod hemocyanin offers a possible mechanism for cooperativity
Biochemistry, 42, 2003
|
|
2W5W
 
 | | Structure of TAB5 alkaline phosphatase mutant His 135 Asp with Zn bound in the M3 site. | | Descriptor: | ALKALINE PHOSPHATASE, ZINC ION | | Authors: | Koutsioulis, D, Lyskowski, A, Maki, S, Guthrie, E, Feller, G, Bouriotis, V, Heikinheimo, P. | | Deposit date: | 2008-12-15 | | Release date: | 2009-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Coordination Sphere of the Third Metal Site is Essential to the Activity and Metal Selectivity of Alkaline Phosphatases.
Protein Sci., 19, 2010
|
|
2W5V
 
 | | Structure of TAB5 alkaline phosphatase mutant His 135 Asp with Mg bound in the M3 site. | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, ZINC ION | | Authors: | Koutsioulis, D, Lyskowski, A, Maki, S, Guthrie, E, Feller, G, Bouriotis, V, Heikinheimo, P. | | Deposit date: | 2008-12-15 | | Release date: | 2009-11-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Coordination Sphere of the Third Metal Site is Essential to the Activity and Metal Selectivity of Alkaline Phosphatases.
Protein Sci., 19, 2010
|
|
2W5X
 
 | | Structure of TAB5 alkaline phosphatase mutant His 135 Glu with Mg bound in the M3 site. | | Descriptor: | ALKALINE PHOSPHATASE, MAGNESIUM ION, ZINC ION | | Authors: | Koutsioulis, D, Lyskowski, A, Maki, S, Guthrie, E, Feller, G, Bouriotis, V, Heikinheimo, P. | | Deposit date: | 2008-12-15 | | Release date: | 2009-11-24 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Coordination Sphere of the Third Metal Site is Essential to the Activity and Metal Selectivity of Alkaline Phosphatases.
Protein Sci., 19, 2010
|
|
