4WOZ
 
 | | Crystal Structures of CdNal from Clostridium difficile in complex with mannosamine | | Descriptor: | 2-(ACETYLAMINO)-2-DEOXY-D-MANNOSE, N-acetylneuraminate lyase | | Authors: | Liu, W.D, Guo, R.T, Cui, Y.F, Chen, X, Wu, Q.Q, Zhu, D.M. | | Deposit date: | 2014-10-17 | | Release date: | 2015-10-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Crystal Structures of CdNal from Clostridium difficile in complex with mannosamine
to be published
|
|
4XLY
 
 | | The complex structure of KS-D75C with substrate CPP | | Descriptor: | (2E)-3-methyl-5-[(1R,4aR,8aR)-5,5,8a-trimethyl-2-methylidenedecahydronaphthalen-1-yl]pent-2-en-1-yl trihydrogen diphosphate, Uncharacterized protein blr2150 | | Authors: | Hu, Y, Zheng, Y, Ko, T.P, Liu, W, Guo, R.T. | | Deposit date: | 2015-01-14 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure, function and inhibition of ent-kaurene synthase from Bradyrhizobium japonicum.
Sci Rep, 4, 2014
|
|
4XLX
 
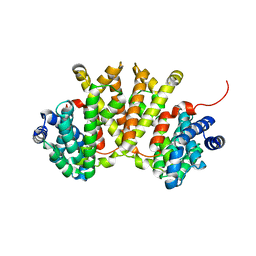 | | Crystal structure of BjKS from Bradyrhizobium japonicum | | Descriptor: | Uncharacterized protein blr2150 | | Authors: | Hu, Y, Zheng, Y, Ko, T.P, Liu, W, Guo, R.T. | | Deposit date: | 2015-01-14 | | Release date: | 2015-02-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure, function and inhibition of ent-kaurene synthase from Bradyrhizobium japonicum.
Sci Rep, 4, 2014
|
|
2ZB4
 
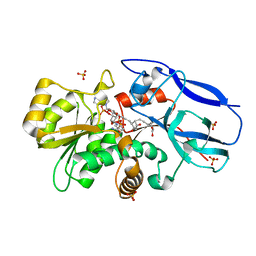 | | Crystal structure of human 15-ketoprostaglandin delta-13-reductase in complex with NADP and 15-keto-PGE2 | | Descriptor: | (5E,13E)-11-HYDROXY-9,15-DIOXOPROSTA-5,13-DIEN-1-OIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prostaglandin reductase 2, ... | | Authors: | Wu, Y.H, Wang, A.H.J, Ko, T.P, Guo, R.T, Hu, S.M, Chuang, L.M. | | Deposit date: | 2007-10-16 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2.
Structure, 16, 2008
|
|
2ZCM
 
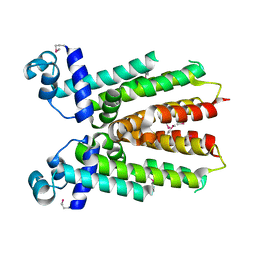 | | Crystal structure of IcaR, a repressor of the TetR family | | Descriptor: | Biofilm operon icaABCD HTH-type negative transcriptional regulator icaR | | Authors: | Jeng, W.Y, Ko, T.P, Liu, C.I, Guo, R.T, Shr, H.L, Wang, A.H.J. | | Deposit date: | 2007-11-10 | | Release date: | 2008-02-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | Crystal structure of IcaR, a repressor of the TetR family implicated in biofilm formation in Staphylococcus epidermidis
Nucleic Acids Res., 36, 2008
|
|
2ZB3
 
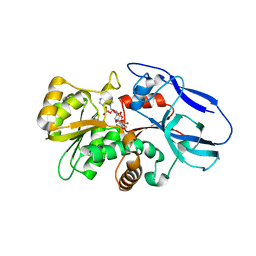 | | Crystal structure of mouse 15-ketoprostaglandin delta-13-reductase in complex with NADPH | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prostaglandin reductase 2 | | Authors: | Wu, Y.H, Wang, A.H.J, Ko, T.P, Guo, R.T, Hu, S.M, Chuang, L.M. | | Deposit date: | 2007-10-16 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2.
Structure, 16, 2008
|
|
2ZCN
 
 | | Crystal structure of IcaR, a repressor of the TetR family | | Descriptor: | Biofilm operon icaABCD HTH-type negative transcriptional regulator icaR | | Authors: | Jeng, W.Y, Ko, T.P, Liu, C.I, Guo, R.T, Liu, C.L, Wang, A.H.J. | | Deposit date: | 2007-11-10 | | Release date: | 2008-02-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of IcaR, a repressor of the TetR family implicated in biofilm formation in Staphylococcus epidermidis
Nucleic Acids Res., 36, 2008
|
|
2ZB8
 
 | | Crystal structure of human 15-ketoprostaglandin delta-13-reductase in complex with NADP and indomethacin | | Descriptor: | INDOMETHACIN, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Prostaglandin reductase 2, ... | | Authors: | Wu, Y.H, Wang, A.H.J, Ko, T.P, Guo, R.T, Hu, S.M, Chuang, L.M. | | Deposit date: | 2007-10-16 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2.
Structure, 16, 2008
|
|
2ZB7
 
 | | Crystal structure of human 15-ketoprostaglandin delta-13-reductase in complex with NADPH and nicotinamide | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NICOTINAMIDE, Prostaglandin reductase 2 | | Authors: | Wu, Y.H, Wang, A.H.J, Ko, T.P, Guo, R.T, Hu, S.M, Chuang, L.M. | | Deposit date: | 2007-10-16 | | Release date: | 2008-09-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for catalytic and inhibitory mechanisms of human prostaglandin reductase PTGR2.
Structure, 16, 2008
|
|
2ZYS
 
 | | A. Fulgidus lipase with fatty acid fragment and chloride | | Descriptor: | CHLORIDE ION, Lipase, putative, ... | | Authors: | Chen, C.K, Ko, T.P, Guo, R.T, Wang, A.H. | | Deposit date: | 2009-01-29 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure of the alkalohyperthermophilic Archaeoglobus fulgidus lipase contains a unique C-terminal domain essential for long-chain substrate binding.
J.Mol.Biol., 390, 2009
|
|
2ZYI
 
 | | A. Fulgidus lipase with fatty acid fragment and calcium | | Descriptor: | CALCIUM ION, Lipase, putative, ... | | Authors: | Chen, C.K, Ko, T.P, Guo, R.T, Wang, A.H. | | Deposit date: | 2009-01-22 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of the alkalohyperthermophilic Archaeoglobus fulgidus lipase contains a unique C-terminal domain essential for long-chain substrate binding.
J.Mol.Biol., 390, 2009
|
|
2ZYH
 
 | | mutant A. Fulgidus lipase S136A complexed with fatty acid fragment | | Descriptor: | CALCIUM ION, HEXADECANE, Lipase, ... | | Authors: | Chen, C.K, Ko, T.P, Guo, R.T, Wang, A.H. | | Deposit date: | 2009-01-22 | | Release date: | 2009-06-16 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure of the alkalohyperthermophilic Archaeoglobus fulgidus lipase contains a unique C-terminal domain essential for long-chain substrate binding.
J.Mol.Biol., 390, 2009
|
|
3AMR
 
 | | Crystal Structures of Bacillus subtilis Alkaline Phytase in Complex with Ca2+, Co2+, Ni2+, Mg2+ and myo-Inositol Hexasulfate | | Descriptor: | 3-phytase, CALCIUM ION, D-MYO-INOSITOL-HEXASULPHATE | | Authors: | Zeng, Y.F, Ko, T.P, Lai, H.L, Cheng, Y.S, Wu, T.H, Ma, Y, Yang, C.S, Cheng, K.J, Huang, C.H, Guo, R.T, Liu, J.R. | | Deposit date: | 2010-08-22 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Crystal structures of Bacillus alkaline phytase in complex with divalent metal ions and inositol hexasulfate
J.Mol.Biol., 409, 2011
|
|
3AMD
 
 | | Crystal structures of Thermotoga maritima Cel5A, apo form and tetramer/au | | Descriptor: | Endoglucanase | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AOF
 
 | | Crystal structures of Thermotoga maritima Cel5A in complex with Mannotriose substrate | | Descriptor: | Endoglucanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-beta-D-mannopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-09-27 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.288 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AMG
 
 | | Crystal structures of Thermotoga maritima Cel5A in complex with Cellobiose substrate, mutant form | | Descriptor: | Endoglucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-20 | | Release date: | 2011-08-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AMS
 
 | | Crystal Structures of Bacillus subtilis Alkaline Phytase in Complex with Ca2+, Cd2+, Co2+, Ni2+, Mg2+ and myo-Inositol Hexasulfate | | Descriptor: | 3-phytase, CADMIUM ION, CALCIUM ION, ... | | Authors: | Zeng, Y.F, Ko, T.P, Lai, H.L, Cheng, Y.S, Wu, T.H, Ma, Y, Yang, C.S, Cheng, K.J, Huang, C.H, Guo, R.T, Liu, J.R. | | Deposit date: | 2010-08-22 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal structures of Bacillus alkaline phytase in complex with divalent metal ions and inositol hexasulfate
J.Mol.Biol., 409, 2011
|
|
3AMC
 
 | | Crystal structures of Thermotoga maritima Cel5A, apo form and dimer/au | | Descriptor: | Endoglucanase | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2010-08-19 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AXE
 
 | | The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase V18Y/W203Y in complex with cellotetraose (cellobiose density was observed) | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucanase, CALCIUM ION, ... | | Authors: | Huang, J.W, Cheng, Y.S, Ko, T.P, Lin, C.Y, Lai, H.L, Chen, C.C, Ma, Y, Huang, C.H, Zheng, Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-04-04 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Rational design to improve thermostability and specific activity of the truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase
Appl.Microbiol.Biotechnol., 94, 2012
|
|
3AXD
 
 | | The truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase V18Y/W203Y in apo-form | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Beta-glucanase, CALCIUM ION | | Authors: | Huang, J.W, Cheng, Y.S, Ko, T.P, Lin, C.Y, Lai, H.L, Chen, C.C, Ma, Y, Huang, C.H, Zheng, Y, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-04-03 | | Release date: | 2012-02-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Rational design to improve thermostability and specific activity of the truncated Fibrobacter succinogenes 1,3-1,4-beta-D-glucanase
Appl.Microbiol.Biotechnol., 94, 2012
|
|
2ZYR
 
 | | A. Fulgidus lipase with fatty acid fragment and magnesium | | Descriptor: | Lipase, putative, MAGNESIUM ION, ... | | Authors: | Chen, C.K, Ko, T.P, Guo, R.T, Wang, A.H. | | Deposit date: | 2009-01-28 | | Release date: | 2009-06-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure of the alkalohyperthermophilic Archaeoglobus fulgidus lipase contains a unique C-terminal domain essential for long-chain substrate binding.
J.Mol.Biol., 390, 2009
|
|
3AZR
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellobiose | | Descriptor: | Endoglucanase, beta-D-glucopyranose, beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AZS
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Mannotriose | | Descriptor: | Endoglucanase, beta-D-mannopyranose-(1-4)-beta-D-mannopyranose-(1-4)-alpha-D-mannopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
3AZT
 
 | | Diverse Substrates Recognition Mechanism Revealed by Thermotoga maritima Cel5A Structures in Complex with Cellotetraose | | Descriptor: | Endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Wu, T.H, Huang, C.H, Ko, T.P, Lai, H.L, Ma, Y, Chen, C.C, Cheng, Y.S, Liu, J.R, Guo, R.T. | | Deposit date: | 2011-05-30 | | Release date: | 2011-08-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Diverse substrate recognition mechanism revealed by Thermotoga maritima Cel5A structures in complex with cellotetraose, cellobiose and mannotriose
Biochim.Biophys.Acta, 1814, 2011
|
|
4TSR
 
 | |
