7JPT
 
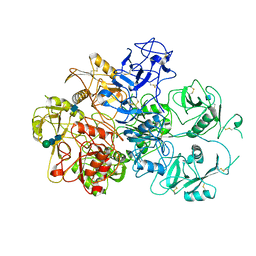 | | Structure of an endocytic receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Lymphocyte antigen 75, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Gully, B.S, Rossjohn, J, Berry, R. | | Deposit date: | 2020-08-09 | | Release date: | 2020-12-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | The cryo-EM structure of the endocytic receptor DEC-205.
J.Biol.Chem., 296, 2020
|
|
7JPU
 
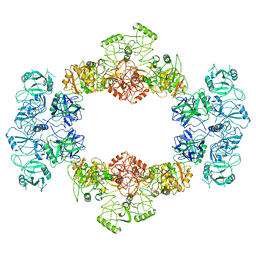 | |
6SIU
 
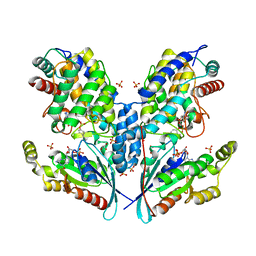 | | Crystal structure of IbpAFic2 covalently tethered to Cdc42 | | Descriptor: | Cell division control protein 42 homolog, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Gulen, B, Roselin, M, Albers, M, Hedberg, C, Itzen, A, Pogenberg, V. | | Deposit date: | 2019-08-12 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Identification of targets of AMPylating Fic enzymes by co-substrate-mediated covalent capture.
Nat.Chem., 12, 2020
|
|
6AVF
 
 | | Crystal structure of the KFJ5 TCR-NY-ESO-1-HLA-B*07:02 complex | | Descriptor: | ALA-PRO-ARG-GLY-PRO-HIS-GLY-GLY-ALA-ALA-SER-GLY-LEU, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Gully, B.S, Gras, S, Rossjohn, J. | | Deposit date: | 2017-09-02 | | Release date: | 2018-02-28 | | Last modified: | 2019-04-17 | | Method: | X-RAY DIFFRACTION (2.028 Å) | | Cite: | Divergent T-cell receptor recognition modes of a HLA-I restricted extended tumour-associated peptide.
Nat Commun, 9, 2018
|
|
6AT5
 
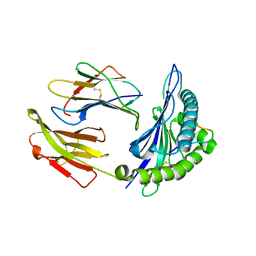 | |
6AVG
 
 | | Crystal structure of the KFJ37 TCR-NY-ESO-1-HLA-B*07:02 complex | | Descriptor: | ALA-PRO-ARG-GLY-PRO-HIS-GLY-GLY-ALA-ALA-SER-GLY-LEU, Beta-2-microglobulin, HLA class I histocompatibility antigen, ... | | Authors: | Gully, B.S, Gras, S, Rossjohn, J. | | Deposit date: | 2017-09-02 | | Release date: | 2018-02-28 | | Last modified: | 2018-03-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Divergent T-cell receptor recognition modes of a HLA-I restricted extended tumour-associated peptide.
Nat Commun, 9, 2018
|
|
6AT6
 
 | | Crystal structure of the KFJ5 TCR | | Descriptor: | T-cell receptor alpha variable 4, T-cell receptor, sp3.4 alpha chain chimera, ... | | Authors: | Gully, B.S, Rossjohn, J. | | Deposit date: | 2017-08-28 | | Release date: | 2018-02-28 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (1.417 Å) | | Cite: | Divergent T-cell receptor recognition modes of a HLA-I restricted extended tumour-associated peptide.
Nat Commun, 9, 2018
|
|
4OZS
 
 | | RNA binding protein | | Descriptor: | Alpha solenoid protein | | Authors: | Gully, B.S, Bond, C.S. | | Deposit date: | 2014-02-19 | | Release date: | 2015-04-22 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | The design and structural characterization of a synthetic pentatricopeptide repeat protein.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
6W9Q
 
 | | Peptide-bound SARS-CoV-2 Nsp9 RNA-replicase | | Descriptor: | 3C-like proteinase peptide, Non-structural protein 9 fusion, PHOSPHATE ION | | Authors: | Littler, D.R, Gully, B.S, Riboldi-Tunnicliffe, A, Rossjohn, J. | | Deposit date: | 2020-03-23 | | Release date: | 2020-04-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of the SARS-CoV-2 Non-structural Protein 9, Nsp9.
Iscience, 23, 2020
|
|
7KRI
 
 | | FR6-bound SARS-CoV-2 Nsp9 RNA-replicase | | Descriptor: | 1,3-dimethyl-1H-pyrrolo[3,4-d]pyrimidine-2,4(3H,6H)-dione, MALONATE ION, Non-structural protein 9, ... | | Authors: | Littler, D.R, Gully, B.S, Rossjohn, J. | | Deposit date: | 2020-11-20 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Binding of a pyrimidine RNA base-mimic to SARS-CoV-2 nonstructural protein 9.
J.Biol.Chem., 297, 2021
|
|
6WXD
 
 | | SARS-CoV-2 Nsp9 RNA-replicase | | Descriptor: | Non-structural protein 9, SULFATE ION | | Authors: | Littler, D.R, Gully, B.S, Riboldi-Tunnicliffe, A, Rossjohn, J. | | Deposit date: | 2020-05-10 | | Release date: | 2020-05-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the SARS-CoV-2 Non-structural Protein 9, Nsp9.
Iscience, 23, 2020
|
|
6WC1
 
 | |
6ZMD
 
 | | Crystal structure of HYPE covalently tethered to BiP bound to AMP-PNP | | Descriptor: | Endoplasmic reticulum chaperone BiP, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Fauser, J, Gulen, B, Pett, C, Hedberg, C, Itzen, A, Pogenberg, V. | | Deposit date: | 2020-07-02 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Specificity of AMPylation of the human chaperone BiP is mediated by TPR motifs of FICD.
Nat Commun, 12, 2021
|
|
7LLI
 
 | | Stimulatory immune receptor protein complex | | Descriptor: | 1-deoxy-1-({2,6-dioxo-5-[(E)-propylideneamino]-1,2,3,6-tetrahydropyrimidin-4-yl}amino)-D-ribitol, Beta-2-microglobulin, Major histocompatibility complex class I-related gene protein, ... | | Authors: | Rice, M.T, Littler, D.R, Rossjohn, J, Gully, B.S. | | Deposit date: | 2021-02-04 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Recognition of the antigen-presenting molecule MR1 by a V delta 3 + gamma delta T cell receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LLJ
 
 | | Inhibitory immune receptor protein complex | | Descriptor: | Beta-2-microglobulin, Major histocompatibility complex class I-related gene protein, N-(6-formyl-4-oxo-3,4-dihydropteridin-2-yl)acetamide, ... | | Authors: | Rice, M.T, Littler, D.R, Rossjohn, J, Gully, B.S. | | Deposit date: | 2021-02-04 | | Release date: | 2021-11-17 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Recognition of the antigen-presenting molecule MR1 by a V delta 3 + gamma delta T cell receptor.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
8DQU
 
 | | Nanobody bound SARS-CoV-2 Nsp9 | | Descriptor: | Nanobody, Non-structural protein 9 | | Authors: | Littler, D.R, Gully, B.S, Rossjohn, J. | | Deposit date: | 2022-07-20 | | Release date: | 2023-02-08 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.45003176 Å) | | Cite: | Inside-out: Antibody-binding reveals potential folding hinge-points within the SARS-CoV-2 replication co-factor nsp9.
Plos One, 18, 2023
|
|
7N3K
 
 | | Oridonin-bound SARS-CoV-2 Nsp9 | | Descriptor: | (1beta,6beta,7beta,8alpha,9beta,10alpha,13alpha,14R,16beta)-1,6,7,14-tetrahydroxy-7,20-epoxykauran-15-one, Non-structural protein 9, SULFATE ION | | Authors: | Littler, D.R, Gully, B.S, Rossjohn, J. | | Deposit date: | 2021-06-01 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A natural product compound inhibits coronaviral replication in vitro by binding to the conserved Nsp9 SARS-CoV-2 protein.
J.Biol.Chem., 297, 2021
|
|
5V52
 
 | | Structure of TIGIT bound to nectin-2 (CD112) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, Nectin-2, ... | | Authors: | Deuss, F.A, Gully, B.S, Rossjohn, J, Berry, R. | | Deposit date: | 2017-03-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Recognition of nectin-2 by the natural killer cell receptor T cell immunoglobulin and ITIM domain (TIGIT).
J. Biol. Chem., 292, 2017
|
|
