1S9Y
 
 | | Crystal Structure Analysis of NY-ESO-1 epitope analogue, SLLMWITQS, in complex with HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Webb, A.I, Dunstone, M.A, Chen, W, Aguilar, M.I, Chen, Q, Chang, L, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Rossjohn, J, Purcell, A.W. | | Deposit date: | 2004-02-05 | | Release date: | 2004-09-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue
J.Biol.Chem., 279, 2004
|
|
1S9W
 
 | | Crystal Structure Analysis of NY-ESO-1 epitope, SLLMWITQC, in complex with HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Webb, A.I, Dunstone, M.A, Chen, W, Aguilar, M.I, Chen, Q, Chang, L, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Rossjohn, J, Purcell, A.W. | | Deposit date: | 2004-02-05 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue
J.Biol.Chem., 279, 2004
|
|
1TZ5
 
 | | [pNPY19-23]-hPP bound to DPC Micelles | | Descriptor: | Pancreatic prohormone,neuropeptide Y,Pancreatic prohormone | | Authors: | Lerch, M, Kamimori, H, Folkers, G, Aguilar, M.I, Beck-Sickinger, A.G, Zerbe, O. | | Deposit date: | 2004-07-09 | | Release date: | 2005-07-05 | | Last modified: | 2020-01-01 | | Method: | SOLUTION NMR | | Cite: | Strongly Altered Receptor Binding Properties in PP and NPY Chimeras Are Accompanied by Changes
in Structure and Membrane Binding
Biochemistry, 44, 2005
|
|
3R87
 
 | |
2HPU
 
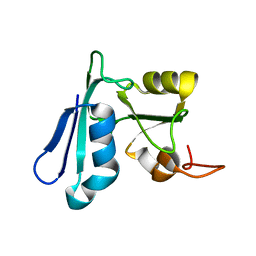 | | Solution NMR structure of the apo-NosL protein from Achromobacter cycloclastes | | Descriptor: | NosL protein | | Authors: | Taubner, L.M, McGuirl, M.A, Dooley, D.M, Copie, V. | | Deposit date: | 2006-07-17 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Studies of Apo Nosl, an Accessory Protein of the Nitrous Oxide Reductase System: Insights from Structural Homology with MerB, a Mercury Resistance Protein.
Biochemistry, 45, 2006
|
|
2HQ3
 
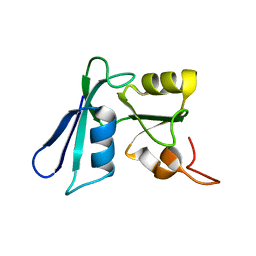 | | Solution NMR structure of the apo-NosL protein from Achromobacter cycloclastes | | Descriptor: | NosL protein | | Authors: | Taubner, L.M, McGuirl, M.A, Dooley, D.M, Copie, V. | | Deposit date: | 2006-07-18 | | Release date: | 2006-10-24 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural Studies of Apo Nosl, an Accessory Protein of the Nitrous Oxide Reductase System: Insights from Structural Homology with MerB, a Mercury Resistance Protein.
Biochemistry, 45, 2006
|
|
2NDA
 
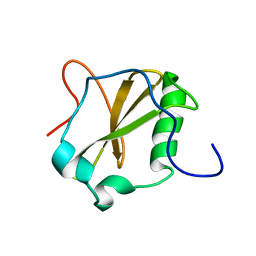 | | Solution structure of MapZ extracellular domain second subdomain | | Descriptor: | Mid-cell-anchored protein Z | | Authors: | Jean, N.L, Manuse, S, Guinot, M, Bougault, C.M, Grangeasse, C, Simorre, J.-P. | | Deposit date: | 2016-05-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of the extracellular domain of the pneumococcal cell division site positioning protein MapZ.
Nat Commun, 7, 2016
|
|
2ND9
 
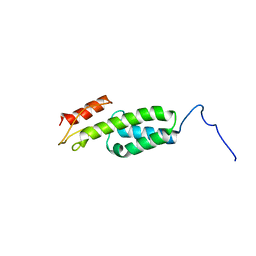 | | Solution structure of MapZ extracellular domain first subdomain | | Descriptor: | Mid-cell-anchored protein Z | | Authors: | Jean, N.L, Manuse, S, Guinot, M, Bougault, C.M, Grangeasse, C, Simorre, J.-P. | | Deposit date: | 2016-05-11 | | Release date: | 2016-06-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-function analysis of the extracellular domain of the pneumococcal cell division site positioning protein MapZ.
Nat Commun, 7, 2016
|
|
2MH2
 
 | | Structural insights into the DNA recognition and protein interaction domains reveal fundamental homologous DNA pairing properties of HOP2 | | Descriptor: | Homologous-pairing protein 2 homolog | | Authors: | Moktan, H, Guiraldelli, M.F, Eyter, C.A, Zhao, W, Camerini-Otero, R.D, Sung, P, Zhou, D.H, Pezza, R.J. | | Deposit date: | 2013-11-13 | | Release date: | 2014-04-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and DNA-binding Properties of the Winged Helix Domain of the Meiotic Recombination HOP2 Protein.
J.Biol.Chem., 289, 2014
|
|
4GWA
 
 | | Crystal Structure of a GH7 Family Cellobiohydrolase from Limnoria quadripunctata | | Descriptor: | GH7 family protein, MAGNESIUM ION | | Authors: | McGeehan, J.E, Martin, R.N.A, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2012-09-01 | | Release date: | 2013-06-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HAQ
 
 | | Crystal Structure of a GH7 family cellobiohydrolase from Limnoria quadripunctata in complex with cellobiose and cellotriose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GH7 family protein, ... | | Authors: | Martin, R.N.A, McGeehan, J.E, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2012-09-27 | | Release date: | 2013-06-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HAP
 
 | | Crystal Structure of a GH7 family cellobiohydrolase from Limnoria quadripunctata in complex with cellobiose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, GH7 family protein, ... | | Authors: | Martin, R.N.A, McGeehan, J.E, Streeter, S.D, Cragg, S.M, Guille, M.J, Schnorr, K.M, Kern, M, Bruce, N.C, McQueen-Mason, S.J. | | Deposit date: | 2012-09-27 | | Release date: | 2013-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural characterization of a unique marine animal family 7 cellobiohydrolase suggests a mechanism of cellulase salt tolerance
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
