2V8N
 
 | | Wild-type Structure of Lactose Permease | | Descriptor: | LACTOSE PERMEASE | | Authors: | Guan, L, Mirza, O, Verner, G, Iwata, S, Kaback, H.R. | | Deposit date: | 2007-08-09 | | Release date: | 2007-09-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural Determination of Wild-Type Lactose Permease.
Proc.Natl.Acad.Sci.USA, 104, 2007
|
|
7L16
 
 | | Crystal structure of sugar-bound melibiose permease MelB | | Descriptor: | Melibiose carrier protein, dodecyl 6-O-alpha-D-galactopyranosyl-beta-D-glucopyranoside | | Authors: | Guan, L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | X-ray crystallography reveals molecular recognition mechanism for sugar binding in a melibiose transporter MelB.
Commun Biol, 4, 2021
|
|
7L17
 
 | | Crystal structure of sugar-bound melibiose permease MelB | | Descriptor: | 4-nitrophenyl alpha-D-galactopyranoside, Melibiose carrier protein | | Authors: | Guan, L. | | Deposit date: | 2020-12-14 | | Release date: | 2021-08-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | X-ray crystallography reveals molecular recognition mechanism for sugar binding in a melibiose transporter MelB.
Commun Biol, 4, 2021
|
|
8T60
 
 | |
8FRH
 
 | | Apo structure of D59C mutant of a melibiose transporter | | Descriptor: | Melibiose permease, PENTAETHYLENE GLYCOL | | Authors: | Guan, L, Hariharan, P. | | Deposit date: | 2023-01-06 | | Release date: | 2024-06-05 | | Last modified: | 2024-12-18 | | Method: | X-RAY DIFFRACTION (3.18 Å) | | Cite: | Distinct roles of the major binding residues in the cation-binding pocket of the melibiose transporter MelB.
J.Biol.Chem., 300, 2024
|
|
8FQ9
 
 | |
6PA0
 
 | | Structure of the G77A mutant in Sodium Chloride | | Descriptor: | Antibody HEAVY fragment, Antibody LIGHT fragment, DIACYL GLYCEROL, ... | | Authors: | Cuello, L.G, Guan, L. | | Deposit date: | 2019-06-11 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure, function, and ion-binding properties of a K+channel stabilized in the 2,4-ion-bound configuration.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2Y5Y
 
 | | Crystal structure of LacY in complex with an affinity inactivator | | Descriptor: | 2-sulfanylethyl beta-D-galactopyranoside, BARIUM ION, LACTOSE PERMEASE | | Authors: | Chaptal, V, Kwon, S, Sawaya, M.R, Guan, L, Kaback, H.R, Abramson, J. | | Deposit date: | 2011-01-19 | | Release date: | 2011-06-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Crystal Structure of Lactose Permease in Complex with an Affinity Inactivator Yields Unique Insight Into Sugar Recognition.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6NFV
 
 | | Structure of the KcsA-G77C mutant or the 2,4-ion bound configuration of a K+ channel selectivity filter. | | Descriptor: | (1S)-2-HYDROXY-1-[(NONANOYLOXY)METHYL]ETHYL MYRISTATE, NONAN-1-OL, POTASSIUM ION, ... | | Authors: | Tilegenova, C, Cortes, D.M, Jahovic, N, Hardy, E, Parameswaran, H, Guan, L, Cuello, L.G. | | Deposit date: | 2018-12-20 | | Release date: | 2019-08-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure, function, and ion-binding properties of a K+channel stabilized in the 2,4-ion-bound configuration.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6NFU
 
 | | Structure of the KcsA-G77A mutant or the 2,4-ion bound configuration of a K+ channel selectivity filter. | | Descriptor: | (1S)-2-HYDROXY-1-[(NONANOYLOXY)METHYL]ETHYL MYRISTATE, NONAN-1-OL, POTASSIUM ION, ... | | Authors: | Tilegenova, C, Cortes, D.M, Jahovic, N, Hardy, E, Parameswaran, H, Guan, L, Cuello, L.G. | | Deposit date: | 2018-12-20 | | Release date: | 2019-08-07 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure, function, and ion-binding properties of a K+channel stabilized in the 2,4-ion-bound configuration.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2CFP
 
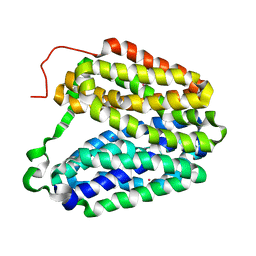 | | Sugar Free Lactose Permease at acidic pH | | Descriptor: | LACTOSE PERMEASE, MERCURY (II) ION | | Authors: | Mirza, O, Guan, L, Verner, G, Iwata, S, Kaback, H.R. | | Deposit date: | 2006-02-22 | | Release date: | 2006-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Evidence for Induced Fit and a Mechanism for Sugar/H(+) Symport in Lacy.
Embo J., 25, 2006
|
|
2CFQ
 
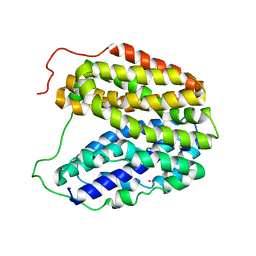 | | Sugar Free Lactose Permease at neutral pH | | Descriptor: | LACTOSE PERMEASE, MERCURY (II) ION | | Authors: | Mirza, O, Guan, L, Verner, G, Iwata, S, Kaback, H.R. | | Deposit date: | 2006-02-22 | | Release date: | 2006-03-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Evidence for Induced Fit and a Mechanism for Sugar/H(+) Symport in Lacy.
Embo J., 25, 2006
|
|
4M64
 
 | |
8XW5
 
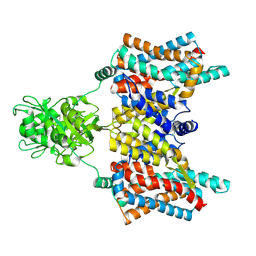 | | Cryo-EM structure of the aspartate:alanine antiporter AspT mutant L60C | | Descriptor: | Aspartate/alanine antiporter | | Authors: | Nanatani, K, Kanno, R, Kawabata, T, Watanabe, S, Hidaka, M, Yamanaka, T, Toda, K, Fujiki, T, Kunii, K, Miyamoto, A, Chiba, F, Ogasawara, S, Murata, T, Humbel, B.M, Inaba, K, Mitsuoka, K, Guan, L, Abe, K, Yamamoto, M, Koshiba, S. | | Deposit date: | 2024-01-16 | | Release date: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.56 Å) | | Cite: | Cryo-EM structure and molecular mechanism of the aspartate:alanine antiporter AspT from Tetragenococcus halophilus
To Be Published
|
|
8Y1X
 
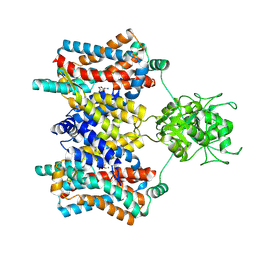 | | Cryo-EM structure of the aspartate:alanine antiporter AspT WT | | Descriptor: | ASPARTIC ACID, Aspartate/alanine antiporter | | Authors: | Nanatani, K, Kanno, R, Kawabata, T, Watanabe, S, Hidaka, M, Yamanaka, T, Toda, K, Fujiki, T, Kunii, K, Miyamoto, A, Chiba, F, Ogasawara, S, Murata, T, Humbel, B.M, Inaba, K, Mitsuoka, K, Guan, L, Abe, K, Yamamoto, M, Koshiba, S. | | Deposit date: | 2024-01-25 | | Release date: | 2025-01-29 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Cryo-EM structure and molecular mechanism of the aspartate:alanine antiporter AspT from Tetragenococcus halophilus
To Be Published
|
|
4O60
 
 | | Structure of ankyrin repeat protein | | Descriptor: | ANK-N5C-317 | | Authors: | Ethayathulla, A.S, Guan, L. | | Deposit date: | 2013-12-20 | | Release date: | 2015-03-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | A transcription blocker isolated from a designed repeat protein combinatorial library by in vivo functional screen.
Sci Rep, 5, 2015
|
|
4QFV
 
 | |
5XHQ
 
 | | Apolipoprotein N-acyl Transferase | | Descriptor: | Apolipoprotein N-acyltransferase, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Yingzhi, X, Yong, X, Guangyuan, L, Fei, S. | | Deposit date: | 2017-04-23 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.587 Å) | | Cite: | Crystal structure of E. coli apolipoprotein N-acyl transferase
Nat Commun, 8, 2017
|
|
9CUO
 
 | | Crystal structure of CRBN with compound 3 | | Descriptor: | (3S)-3-(3-methyl-2-oxo-2,3-dihydro-1H-1,3-benzimidazol-1-yl)piperidine-2,6-dione, 1,2-ETHANEDIOL, Protein cereblon, ... | | Authors: | Zheng, X, Ji, N, Campbell, V, Slavin, A, Zhu, X, Chen, D, Rong, H, Enerson, B, Mayo, M, Sharma, K, Browne, C.M, Klaus, C.R, Li, H, Massa, G, McDonald, A.A, Shi, Y, Sintchak, M, Skouras, S, Walther, D.M, Yuan, K, Zhang, Y, Kelleher, J, Guang, L, Luo, X, Mainolfi, N, Weiss, M.M. | | Deposit date: | 2024-07-26 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of KT-474─a Potent, Selective, and Orally Bioavailable IRAK4 Degrader for the Treatment of Autoimmune Diseases.
J.Med.Chem., 67, 2024
|
|
