4R7J
 
 | | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase with the Internal Deletion Containing CBS Domain from Campylobacter jejuni | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Makowska-Grzyska, M, Gu, M, Hedstrom, L, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2014-08-27 | | Release date: | 2014-09-17 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1172 Å) | | Cite: | Crystal Structure of Inosine 5'-monophosphate Dehydrogenase with the Internal Deletion Containing CBS Domain from Campylobacter jejuni
To be Published, 2014
|
|
3KWO
 
 | | Crystal Structure of Putative Bacterioferritin from Campylobacter jejuni | | Descriptor: | 1,4-BUTANEDIOL, ACETIC ACID, GLYCEROL, ... | | Authors: | Kim, Y, Gu, M, Papazisi, L, Anderson, W, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-12-01 | | Release date: | 2010-01-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.985 Å) | | Cite: | Crystal Structure of Putative Bacterioferritin from Campylobacter jejuni
To be Published
|
|
3LNO
 
 | | Crystal Structure of Domain of Unknown Function DUF59 from Bacillus anthracis | | Descriptor: | Putative uncharacterized protein | | Authors: | Kim, Y, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-02-02 | | Release date: | 2010-02-16 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Domain of Unknown Function DUF59 Bacillus anthracis
To be Published
|
|
7VFJ
 
 | | Cytochrome c-type biogenesis protein CcmABCD | | Descriptor: | Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, Heme exporter protein C, ... | | Authors: | Zhu, J.P, Zhang, K, Li, J, Zheng, W, Gu, M. | | Deposit date: | 2021-09-13 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.98 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7VFP
 
 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with heme and single ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-09-13 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
6NRU
 
 | | Crystal Structure of the Alpha-ribazole Phosphatase from Shigella flexneri | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CobC, ... | | Authors: | Kim, Y, Gu, M, Shatsman, S, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2019-01-24 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.505 Å) | | Cite: | Crystal Structure of the Alpha-ribazole Phosphatase from Shigella flexneri
To Be Published
|
|
3M5U
 
 | | Crystal Structure of Phosphoserine Aminotransferase from Campylobacter jejuni | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, Phosphoserine aminotransferase | | Authors: | Kim, Y, Gu, M, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-03-13 | | Release date: | 2010-04-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Crystal Structure of Phosphoserine Aminotransferase from Campylobacter jejuni
To be Published
|
|
3MY7
 
 | | The Crystal Structure of the ACDH domain of an Alcohol Dehydrogenase from Vibrio parahaemolyticus to 2.25A | | Descriptor: | Alcohol dehydrogenase/acetaldehyde dehydrogenase, CHLORIDE ION | | Authors: | Stein, A.J, Weger, A, Volkart, L, Gu, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-05-10 | | Release date: | 2010-06-02 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of the ACDH domain of an Alcohol Dehydrogenase from Vibrio parahaemolyticus to 2.25A
To be Published
|
|
3NEU
 
 | |
3LJL
 
 | |
3NTX
 
 | | Crystal Structure of L-asparaginase I from Yersinia pestis | | Descriptor: | Cytoplasmic L-asparaginase I, GLYCEROL | | Authors: | Kim, Y, Gu, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-07-05 | | Release date: | 2010-08-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of L-asparaginase I from Yersinia pestis
To be Published, 2010
|
|
3NWG
 
 | | The crystal structure of a microcomparments protein from Desulfitobacterium hafniense DCB | | Descriptor: | GLYCEROL, Microcompartments protein | | Authors: | Fan, Y, Volkart, L, Gu, M, Axen, S, Greenleaf, W.B, Kerfeld, C, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2010-07-09 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of a PduT homolog from a novel bacterial microcompartment
To be Published
|
|
4ESE
 
 | | The crystal structure of azoreductase from Yersinia pestis CO92 in complex with FMN. | | Descriptor: | DODECAETHYLENE GLYCOL, FLAVIN MONONUCLEOTIDE, FMN-dependent NADH-azoreductase, ... | | Authors: | Tan, K, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-23 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The crystal structure of acyl carrier protein phosphodiesterase from Yersinia pestis CO92 in complex with FMN.
To be Published
|
|
3U1B
 
 | | Crystal structure of the S238R mutant of mycrocine immunity protein (MccF) with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Microcin immunity protein MccF | | Authors: | Nocek, B, Gu, M, Zhou, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-29 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.604 Å) | | Cite: | Structural and Functional Characterization of Microcin C Resistance Peptidase MccF from Bacillus anthracis.
J.Mol.Biol., 420, 2012
|
|
4GCO
 
 | | Central domain of stress-induced protein-1 (STI-1) from C.elegans | | Descriptor: | Protein STI-1 | | Authors: | Osipiuk, J, Bigelow, L, Gu, M, Van Oosten-Hawle, P, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-07-30 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Central domain of stress-induced protein-1 (STI-1) from C.elegans
To be Published
|
|
4GCN
 
 | | N-terminal domain of stress-induced protein-1 (STI-1) from C.elegans | | Descriptor: | Protein STI-1, TRIETHYLENE GLYCOL | | Authors: | Osipiuk, J, Bigelow, L, Gu, M, Van Oosten-Hawle, P, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-07-30 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | N-terminal domain of stress-induced protein-1 (STI-1) from C.elegans
To be Published
|
|
3SR3
 
 | | Crystal structure of the w180a mutant of microcin immunity protein mccf from Bacillus anthracis shows the active site loop in the open conformation. | | Descriptor: | Microcin immunity protein MccF | | Authors: | Nocek, B, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-07-06 | | Release date: | 2011-08-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.495 Å) | | Cite: | Structural and functional characterization of microcin C resistance peptidase MccF from Bacillus anthracis.
J.Mol.Biol., 420, 2012
|
|
4GJ1
 
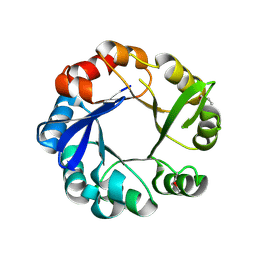 | | Crystal structure of 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase (hisA). | | Descriptor: | 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase | | Authors: | Nocek, B, Gu, M, Kwon, K, Anderson, W.F, Joachimiak, A, CSGID, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-09 | | Release date: | 2012-08-22 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.152 Å) | | Cite: | Crystal structure of 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino] imidazole-4-carboxamide isomerase (hisA).
To be Published
|
|
7F02
 
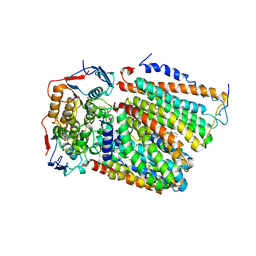 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
7F03
 
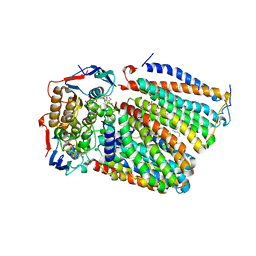 | | Cytochrome c-type biogenesis protein CcmABCD from E. coli in complex with ANP | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, Cytochrome c biogenesis ATP-binding export protein CcmA, Heme exporter protein B, ... | | Authors: | Li, J, Zheng, W, Gu, M, Zhang, K, Zhu, J.P. | | Deposit date: | 2021-06-03 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | Structures of the CcmABCD heme release complex at multiple states.
Nat Commun, 13, 2022
|
|
4EH1
 
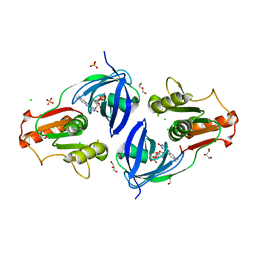 | | Crystal Structure of the Flavohem-like-FAD/NAD Binding Domain of Nitric Oxide Dioxygenase from Vibrio cholerae O1 biovar El Tor | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, Flavohemoprotein, ... | | Authors: | Kim, Y, Gu, M, Hasseman, J, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-02 | | Release date: | 2012-04-25 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of the Flavohem-like-FAD/NAD Binding Domain of Nitric Oxide Dioxygenase from Vibrio cholerae O1 biovar El Tor
To be Published
|
|
3UXV
 
 | | Crystal Structure of 7-cyano-7-deazaguanine reductase, QueF from Vibrio cholerae complexed with NADP and PreQ | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, GUANINE, ... | | Authors: | Kim, Y, Zhou, M, Gu, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-12-05 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Crystal Structure of 7-cyano-7-deazaguanine reductase, QueF from Vibrio cholerae complexed with NADP and PreQ
To be Published, 2012
|
|
3TYX
 
 | | Crystal structure of the F177S mutant of mycrocine immunity protein (MccF) with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Microcin immunity protein MccF | | Authors: | Nocek, B, Gu, M, Zhou, M, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2011-09-26 | | Release date: | 2011-12-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural and Functional Characterization of Microcin C Resistance Peptidase MccF from Bacillus anthracis.
J.Mol.Biol., 420, 2012
|
|
2O5H
 
 | | Uncharacterized Protein Conserved in Bacteria, COG3792 from Neisseria meningitidis | | Descriptor: | Hypothetical protein | | Authors: | Kim, Y, Li, H, Gu, M, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-06 | | Release date: | 2007-01-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Uncharacterized Protein Conserved in Bacteria, COG3792 from Neisseria meningitidis
To be Published
|
|
4GQT
 
 | | N-terminal domain of C. elegans Hsp90 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Heat shock protein 90, ZINC ION | | Authors: | Osipiuk, J, Chhor, G, Gu, M, Van Oosten-Hawle, P, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2012-08-23 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | N-terminal domain of C. elegans Hsp90
To be Published
|
|
